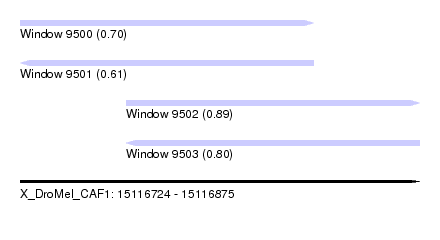
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,116,724 – 15,116,875 |
| Length | 151 |
| Max. P | 0.886377 |

| Location | 15,116,724 – 15,116,835 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.03 |
| Mean single sequence MFE | -35.92 |
| Consensus MFE | -25.35 |
| Energy contribution | -26.73 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15116724 111 + 22224390 UCGCCAAAAUGCCAGAUUGGCUAGGACUCAAAUGUAGCGUAAGCCAAGUGAAGCCCGGAAAAGUUCAUU-CGGAUUUUGGGGAGCUUCUUCAUGAAUUUCAUGAGUUGUCCA-------- ..(((((.........)))))..((((.........((....)).....(((((((...(((((((...-.)))))))..)).)))))((((((.....))))))..)))).-------- ( -32.50) >DroSec_CAF1 28407 111 + 1 UCGCCAAAAUGGCAGAUUGGCUAGGACUCAAAUGUACCGUAAGCCAAGUGAAACCCGGAAAAGUUCAUU-CGGAUUUCGGGGGGCUUCUUCAUGAAUUUCAUGAGUUGCCCA-------- ..(((.....)))...((((((.((((......)).))...))))))......((((((...((((...-.))))))))))((((...((((((.....))))))..)))).-------- ( -39.40) >DroEre_CAF1 35595 109 + 1 UCGCCAAAAUGGCAGAUUGGCUAGGACUCAAAUGUACCGUAAGCCCAGUGAAGCCUGGAAAAGUUCAUU-CGGAUUUCGG--GGCUUCUUCAUGAAUUUCAUGAGCUGUCCA-------- ..(((.....))).....((((.((((......)).))...))))(((((((((((.(((..((((...-.))))))).)--)))))).(((((.....)))))))))....-------- ( -37.10) >DroYak_CAF1 35677 119 + 1 UCGCCAGAAUGCCAGAUUGGCUAGGACUCAAAUGUACCGUAAGCCAAGAGAAGCCUGGAAAAGUUCAUUCCGGAUUUCGG-GGGCUUCUUCAUGAAUUUCAUGAGUUGUCCAAAUGUCCA ......((((.((((.((((((.((((......)).))...)))))).......))))....)))).....((((.....-((((...((((((.....))))))..))))....)))). ( -34.70) >consensus UCGCCAAAAUGCCAGAUUGGCUAGGACUCAAAUGUACCGUAAGCCAAGUGAAGCCCGGAAAAGUUCAUU_CGGAUUUCGG_GGGCUUCUUCAUGAAUUUCAUGAGUUGUCCA________ ..........(((((.((((((.((((......)).))...))))))..(((((((....((((((.....))))))....)))))))((((((.....))))))))))).......... (-25.35 = -26.73 + 1.37)


| Location | 15,116,724 – 15,116,835 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.03 |
| Mean single sequence MFE | -31.12 |
| Consensus MFE | -24.93 |
| Energy contribution | -25.37 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.606339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15116724 111 - 22224390 --------UGGACAACUCAUGAAAUUCAUGAAGAAGCUCCCCAAAAUCCG-AAUGAACUUUUCCGGGCUUCACUUGGCUUACGCUACAUUUGAGUCCUAGCCAAUCUGGCAUUUUGGCGA --------.((((...(((((.....))))).(((((((...((((((..-...))..))))..)))))))...((((....)))).......))))..(((((.........))))).. ( -27.20) >DroSec_CAF1 28407 111 - 1 --------UGGGCAACUCAUGAAAUUCAUGAAGAAGCCCCCCGAAAUCCG-AAUGAACUUUUCCGGGUUUCACUUGGCUUACGGUACAUUUGAGUCCUAGCCAAUCUGCCAUUUUGGCGA --------.((((..((((((.....)))).))..))))...((((((((-............))))))))..((((((...((.((......)))).))))))..((((.....)))). ( -36.60) >DroEre_CAF1 35595 109 - 1 --------UGGACAGCUCAUGAAAUUCAUGAAGAAGCC--CCGAAAUCCG-AAUGAACUUUUCCAGGCUUCACUGGGCUUACGGUACAUUUGAGUCCUAGCCAAUCUGCCAUUUUGGCGA --------.((((((.(((((.....))))).((((((--..((((....-........))))..)))))).)))((((...((.((......)))).))))..)))(((.....))).. ( -31.30) >DroYak_CAF1 35677 119 - 1 UGGACAUUUGGACAACUCAUGAAAUUCAUGAAGAAGCCC-CCGAAAUCCGGAAUGAACUUUUCCAGGCUUCUCUUGGCUUACGGUACAUUUGAGUCCUAGCCAAUCUGGCAUUCUGGCGA ((..(....)..))..(((((.....)))))(((((((.-..(((..(.((((.......)))).)..)))..((((((...((.((......)))).))))))...))).))))..... ( -29.40) >consensus ________UGGACAACUCAUGAAAUUCAUGAAGAAGCCC_CCGAAAUCCG_AAUGAACUUUUCCAGGCUUCACUUGGCUUACGGUACAUUUGAGUCCUAGCCAAUCUGCCAUUUUGGCGA .........((((...(((((.....))))).(((((((...((((.............)))).)))))))......................))))..(((((.........))))).. (-24.93 = -25.37 + 0.44)


| Location | 15,116,764 – 15,116,875 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.04 |
| Mean single sequence MFE | -36.62 |
| Consensus MFE | -28.10 |
| Energy contribution | -29.22 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15116764 111 + 22224390 AAGCCAAGUGAAGCCCGGAAAAGUUCAUU-CGGAUUUUGGGGAGCUUCUUCAUGAAUUUCAUGAGUUGUCCA--------AAUGUCGGCAUCUGUGUCCAGAGCAUUUGGCACUGAUUAU ..((((((((..(((((((........))-))(((((((((.(((....(((((.....)))))))).))))--------)).)))))).((((....)))).))))))))......... ( -36.20) >DroSec_CAF1 28447 111 + 1 AAGCCAAGUGAAACCCGGAAAAGUUCAUU-CGGAUUUCGGGGGGCUUCUUCAUGAAUUUCAUGAGUUGCCCA--------AAUGUCCGCAUCUGUGUCCAGAGCAUUUGGCACUGAUUAU ..((((((((...((((((...((((...-.))))))))))((((...((((((.....))))))..)))).--------..((..(((....)))..))...))))))))......... ( -39.50) >DroEre_CAF1 35635 109 + 1 AAGCCCAGUGAAGCCUGGAAAAGUUCAUU-CGGAUUUCGG--GGCUUCUUCAUGAAUUUCAUGAGCUGUCCA--------AAUGUCCGCAUCUGUGUCCAGAGCAUUUGGCACUGAUUAU .....(((((((((((.(((..((((...-.))))))).)--))))))((((((.....))))))..(.(((--------(((((.....((((....)))))))))))))))))..... ( -35.40) >DroYak_CAF1 35717 119 + 1 AAGCCAAGAGAAGCCUGGAAAAGUUCAUUCCGGAUUUCGG-GGGCUUCUUCAUGAAUUUCAUGAGUUGUCCAAAUGUCCAAAUGUCCGCAUCUGUGUCCAGAGCAUUUGGCACUGAUUAU ..((((((....(((((((...(..((((..((((.....-((((...((((((.....))))))..))))....)))).))))..)((....)).))))).)).))))))......... ( -35.40) >consensus AAGCCAAGUGAAGCCCGGAAAAGUUCAUU_CGGAUUUCGG_GGGCUUCUUCAUGAAUUUCAUGAGUUGUCCA________AAUGUCCGCAUCUGUGUCCAGAGCAUUUGGCACUGAUUAU ..((((((((((((((....((((((.....))))))....)))))))((((((.....))))))......................((.((((....)))))))))))))......... (-28.10 = -29.22 + 1.12)



| Location | 15,116,764 – 15,116,875 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.04 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -24.82 |
| Energy contribution | -25.45 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15116764 111 - 22224390 AUAAUCAGUGCCAAAUGCUCUGGACACAGAUGCCGACAUU--------UGGACAACUCAUGAAAUUCAUGAAGAAGCUCCCCAAAAUCCG-AAUGAACUUUUCCGGGCUUCACUUGGCUU .........(((((.((.((((....)))).(((..((((--------((((....(((((.....))))).(........)....))))-))))..(......))))..)).))))).. ( -25.80) >DroSec_CAF1 28447 111 - 1 AUAAUCAGUGCCAAAUGCUCUGGACACAGAUGCGGACAUU--------UGGGCAACUCAUGAAAUUCAUGAAGAAGCCCCCCGAAAUCCG-AAUGAACUUUUCCGGGUUUCACUUGGCUU .........(((((.(((((((....)))).)))......--------.((((..((((((.....)))).))..))))...((((((((-............))))))))..))))).. ( -35.80) >DroEre_CAF1 35635 109 - 1 AUAAUCAGUGCCAAAUGCUCUGGACACAGAUGCGGACAUU--------UGGACAGCUCAUGAAAUUCAUGAAGAAGCC--CCGAAAUCCG-AAUGAACUUUUCCAGGCUUCACUGGGCUU ....(((((.(((((((.((((.(......).))))))))--------))).....(((((.....))))).((((((--..((((....-........))))..))))))))))).... ( -31.50) >DroYak_CAF1 35717 119 - 1 AUAAUCAGUGCCAAAUGCUCUGGACACAGAUGCGGACAUUUGGACAUUUGGACAACUCAUGAAAUUCAUGAAGAAGCCC-CCGAAAUCCGGAAUGAACUUUUCCAGGCUUCUCUUGGCUU .........(((((.(((((((....)))).))).....(((..(....)..))).(((((.....)))))(((((((.-..(.....)((((.......)))).))))))).))))).. ( -31.80) >consensus AUAAUCAGUGCCAAAUGCUCUGGACACAGAUGCGGACAUU________UGGACAACUCAUGAAAUUCAUGAAGAAGCCC_CCGAAAUCCG_AAUGAACUUUUCCAGGCUUCACUUGGCUU .........(((((..((((((....)))).)).......................(((((.....))))).(((((((...((((.............)))).)))))))..))))).. (-24.82 = -25.45 + 0.63)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:01 2006