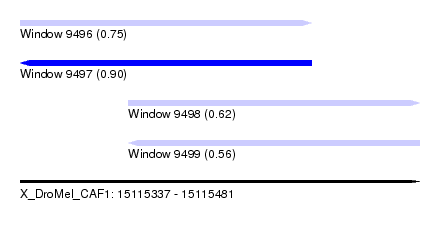
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,115,337 – 15,115,481 |
| Length | 144 |
| Max. P | 0.901861 |

| Location | 15,115,337 – 15,115,442 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.39 |
| Mean single sequence MFE | -31.33 |
| Consensus MFE | -26.64 |
| Energy contribution | -27.32 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15115337 105 + 22224390 AUUUGUUUGGGUUUCGGCCCGGCCU-GUGUGCAUCCACUUGUUACGGUUCAGUUUUUGACAUUCGUCAAAAAUUGUGUCACAAUUUCAGCGAACUUUUCGCCGGCC-------------- ........((((....))))(((((-(((((((......)).)))((..(((((((((((....)))))))))))..)))))......((((.....)))).))))-------------- ( -32.20) >DroSec_CAF1 27054 106 + 1 AUUUGUUUGGGUUUCGGCCCGGCCCGGUGUGCAUCCACUUGUUACGGUUCAGUUUUUGACAUUCGUCAAAAAUUGUGUCACAAUUUCAGCGAACUUUUCGCCGGUC-------------- ......((((((....)))))).(((((((((......((((.(((...(((((((((((....)))))))))))))).)))).....))).......))))))..-------------- ( -29.61) >DroEre_CAF1 34354 104 + 1 AUUUGUUUGGGUUUCGGCCCG-CCU-GUGUGCAUCCACUUGUUACGGUUCAGUUUUUGACAUUCGUCAAAAAUUGUGUCACAAUUUCAGCGAACUUUCCGCCGGCC-------------- ........((((....))))(-(((-(((((((......)).)))((..(((((((((((....)))))))))))..)))))......(((.......))).))).-------------- ( -28.20) >DroYak_CAF1 34290 118 + 1 AUUUGUUUGGGUUUCGGCCCG-CCU-GUGUGCAUCCACUUGUUACGGUUCAGUUUUUGACAUUCGUCAAAAAUUGUGUCACAAUUUCAGCGAACUUUUCGCCGGCCAACGGCCAAUGGCC ........((((....))))(-(((-(((((((......)).)))((..(((((((((((....)))))))))))..)))))......((((.....)))).((((...))))...))). ( -35.30) >consensus AUUUGUUUGGGUUUCGGCCCG_CCU_GUGUGCAUCCACUUGUUACGGUUCAGUUUUUGACAUUCGUCAAAAAUUGUGUCACAAUUUCAGCGAACUUUUCGCCGGCC______________ ........((((....))))((((..(((.(((......))))))((..(((((((((((....)))))))))))..)).........((((.....)))).)))).............. (-26.64 = -27.32 + 0.69)

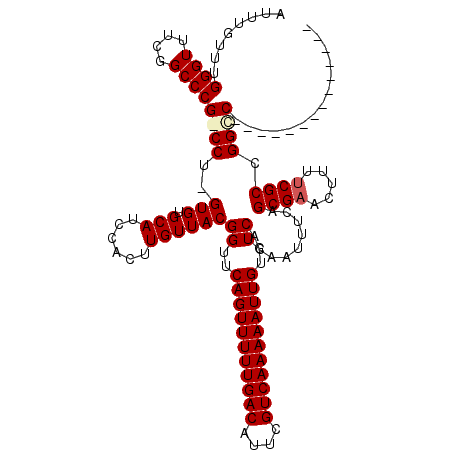

| Location | 15,115,337 – 15,115,442 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.39 |
| Mean single sequence MFE | -31.20 |
| Consensus MFE | -25.04 |
| Energy contribution | -25.10 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15115337 105 - 22224390 --------------GGCCGGCGAAAAGUUCGCUGAAAUUGUGACACAAUUUUUGACGAAUGUCAAAAACUGAACCGUAACAAGUGGAUGCACAC-AGGCCGGGCCGAAACCCAAACAAAU --------------(((((((((.....))))).....((((.((((.((((((((....)))))))).))..(((.......))).)))))).-.))))(((......)))........ ( -31.40) >DroSec_CAF1 27054 106 - 1 --------------GACCGGCGAAAAGUUCGCUGAAAUUGUGACACAAUUUUUGACGAAUGUCAAAAACUGAACCGUAACAAGUGGAUGCACACCGGGCCGGGCCGAAACCCAAACAAAU --------------...((((((...((((((.(((((((.....))))))).).))))).)).....(((..(((......(((......))))))..))))))).............. ( -27.90) >DroEre_CAF1 34354 104 - 1 --------------GGCCGGCGGAAAGUUCGCUGAAAUUGUGACACAAUUUUUGACGAAUGUCAAAAACUGAACCGUAACAAGUGGAUGCACAC-AGG-CGGGCCGAAACCCAAACAAAU --------------((((.((((...((.(((.......))))).((.((((((((....)))))))).))..)).......(((......)))-..)-).))))............... ( -29.60) >DroYak_CAF1 34290 118 - 1 GGCCAUUGGCCGUUGGCCGGCGAAAAGUUCGCUGAAAUUGUGACACAAUUUUUGACGAAUGUCAAAAACUGAACCGUAACAAGUGGAUGCACAC-AGG-CGGGCCGAAACCCAAACAAAU ((((...)))).((((((.((((...((((((.(((((((.....))))))).).))))).))...................(((......)))-..)-).))))))............. ( -35.90) >consensus ______________GGCCGGCGAAAAGUUCGCUGAAAUUGUGACACAAUUUUUGACGAAUGUCAAAAACUGAACCGUAACAAGUGGAUGCACAC_AGG_CGGGCCGAAACCCAAACAAAU ..............((((((((((...)))))).....((((.((((.((((((((....)))))))).))..(((.......))).))))))........))))............... (-25.04 = -25.10 + 0.06)


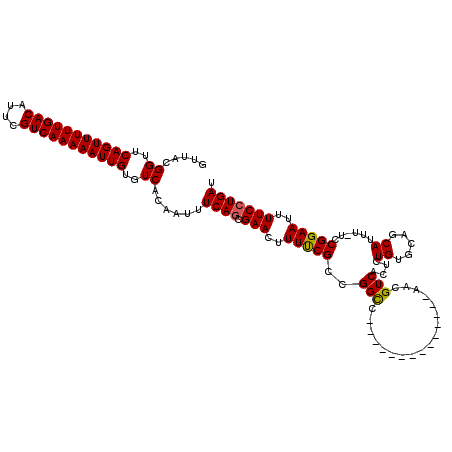
| Location | 15,115,376 – 15,115,481 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -20.75 |
| Energy contribution | -20.62 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15115376 105 + 22224390 GUUACGGUUCAGUUUUUGACAUUCGUCAAAAAUUGUGUCACAAUUUCAGCGAACUUUUCGCCGGCC--------------AAUGUCACUCUGUGCAGCAUUU-UCCGGAAUUUUCGUGAU (((((((..(((((((((((....)))))))))))..))..((((((.((((.....)))).((..--------------((((((((...)))..))))).-.))))))))...))))) ( -28.30) >DroSec_CAF1 27094 105 + 1 GUUACGGUUCAGUUUUUGACAUUCGUCAAAAAUUGUGUCACAAUUUCAGCGAACUUUUCGCCGGUC--------------AACGUCACUCUGUGCAGCAUUU-UCCGGAAUUUUCCUGAU .....((..(((((((((((....)))))))))))..))......((((.(((.......((((..--------------((.(((((...)))..)).)).-.))))....))))))). ( -24.80) >DroEre_CAF1 34392 106 + 1 GUUACGGUUCAGUUUUUGACAUUCGUCAAAAAUUGUGUCACAAUUUCAGCGAACUUUCCGCCGGCC--------------AACGUCACUCUGUGCAGCAUUUUUUCGGAAUUUUCCUGAU .....((..(((((((((((....)))))))))))..))......((((.(((..(((((...((.--------------.(((......)))...)).......)))))..))))))). ( -27.20) >DroYak_CAF1 34328 119 + 1 GUUACGGUUCAGUUUUUGACAUUCGUCAAAAAUUGUGUCACAAUUUCAGCGAACUUUUCGCCGGCCAACGGCCAAUGGCCAACGUCACUCUGUGCAGCAUUU-UCCGGAAUUUUCCUGAU .....((..(((((((((((....)))))))))))..))......((((.(((..(((((..(((((........)))))...(.(((...)))).......-..)))))..))))))). ( -32.80) >consensus GUUACGGUUCAGUUUUUGACAUUCGUCAAAAAUUGUGUCACAAUUUCAGCGAACUUUUCGCCGGCC______________AACGUCACUCUGUGCAGCAUUU_UCCGGAAUUUUCCUGAU .....((..(((((((((((....)))))))))))..))......((((.(((..(((((..(((..................)))....((.....))......)))))..))))))). (-20.75 = -20.62 + -0.13)


| Location | 15,115,376 – 15,115,481 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -28.32 |
| Consensus MFE | -20.95 |
| Energy contribution | -20.82 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559404 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15115376 105 - 22224390 AUCACGAAAAUUCCGGA-AAAUGCUGCACAGAGUGACAUU--------------GGCCGGCGAAAAGUUCGCUGAAAUUGUGACACAAUUUUUGACGAAUGUCAAAAACUGAACCGUAAC .((((((......(((.-.((((..((.....))..))))--------------..)))((((.....)))).....))))))..((.((((((((....)))))))).))......... ( -27.40) >DroSec_CAF1 27094 105 - 1 AUCAGGAAAAUUCCGGA-AAAUGCUGCACAGAGUGACGUU--------------GACCGGCGAAAAGUUCGCUGAAAUUGUGACACAAUUUUUGACGAAUGUCAAAAACUGAACCGUAAC .((((.......((((.-.((((..((.....))..))))--------------..)))).((...((((((.(((((((.....))))))).).))))).)).....))))........ ( -26.20) >DroEre_CAF1 34392 106 - 1 AUCAGGAAAAUUCCGAAAAAAUGCUGCACAGAGUGACGUU--------------GGCCGGCGGAAAGUUCGCUGAAAUUGUGACACAAUUUUUGACGAAUGUCAAAAACUGAACCGUAAC .(((((((..(((((....((((..((.....))..))))--------------......)))))..))).))))..........((.((((((((....)))))))).))......... ( -25.10) >DroYak_CAF1 34328 119 - 1 AUCAGGAAAAUUCCGGA-AAAUGCUGCACAGAGUGACGUUGGCCAUUGGCCGUUGGCCGGCGAAAAGUUCGCUGAAAUUGUGACACAAUUUUUGACGAAUGUCAAAAACUGAACCGUAAC .((.(((....))).))-...(((.(..(((.(((.(((((((((........)))))))))......((((.......)))))))..((((((((....)))))))))))..).))).. ( -34.60) >consensus AUCAGGAAAAUUCCGGA_AAAUGCUGCACAGAGUGACGUU______________GGCCGGCGAAAAGUUCGCUGAAAUUGUGACACAAUUUUUGACGAAUGUCAAAAACUGAACCGUAAC ....(((....))).......(((.(..(((..(((((((.................(((((((...)))))))((((((.....))))))......)))))))....)))..).))).. (-20.95 = -20.82 + -0.13)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:58 2006