
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,114,188 – 15,114,285 |
| Length | 97 |
| Max. P | 0.999157 |

| Location | 15,114,188 – 15,114,285 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 76.96 |
| Mean single sequence MFE | -17.43 |
| Consensus MFE | -9.62 |
| Energy contribution | -10.12 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
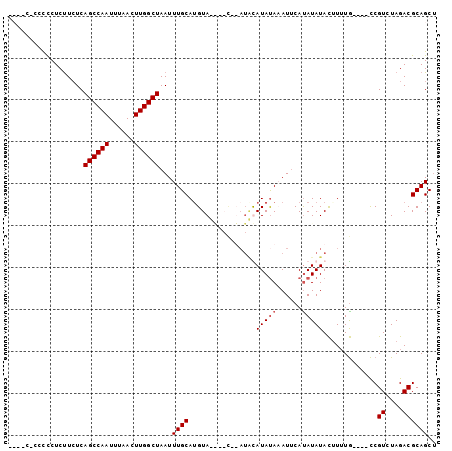
>X_DroMel_CAF1 15114188 97 + 22224390 GCCUCCGCCCCUCCUCUCAGCCAAUUUAACUUGGCUAAUUUGCAUGUACA-AUAUAUGUAUAUAAAUUCAUAUAUACUUUUG----CCGUCUAGACGCAGCU ......((..........((((((......))))))....(((.......-......(((((((......))))))).....----..((....))))))). ( -16.10) >DroSec_CAF1 25914 86 + 1 ------UCUCCUCUUCUCAGCCAAUUUAACUUGGCUAAUUUGCAUGUA----------UAUAUAAAUUCAUAUAUAUAUGUAUACUUCGUCUAGACGCAGCU ------............((((((......))))))....((((((((----------(((((......)))))))))))))..((.(((....))).)).. ( -20.80) >DroEre_CAF1 33080 88 + 1 ---GC-CACUCUCUUCUCAGCCAAUUUAACUUGGCUAAUUUGCAUGUA----C--AUACAUACACAUGCACAUAUACUUUUG----CCGUCUAGACGCAGCC ---..-............((((((......))))))....(((((((.----.--........))))))).........(((----(.((....)))))).. ( -16.90) >DroYak_CAF1 32993 97 + 1 GCCCC-CCCCCUCUUAUCAGCCAAUUUAACUUGGCUAAUUUGCAAGUAUGUACAUAUACAUAUACAUUCAUAUAUACUUUUG----CCGUCUAGACGCAGCU .....-............((((((......)))))).....((..(((((((....))))))).................((----(.((....))))))). ( -15.90) >consensus ____C_CCCCCUCUUCUCAGCCAAUUUAACUUGGCUAAUUUGCAUGUA____C__AUACAUAUAAAUUCAUAUAUACUUUUG____CCGUCUAGACGCAGCU ..................((((((......))))))...((((................(((((......))))).............((....)))))).. ( -9.62 = -10.12 + 0.50)


| Location | 15,114,188 – 15,114,285 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 76.96 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -15.88 |
| Energy contribution | -15.38 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.63 |
| SVM decision value | 3.40 |
| SVM RNA-class probability | 0.999157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
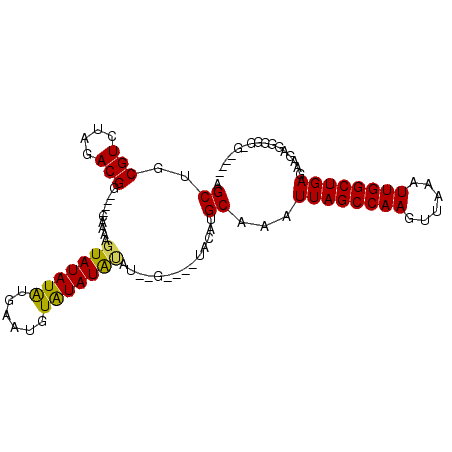
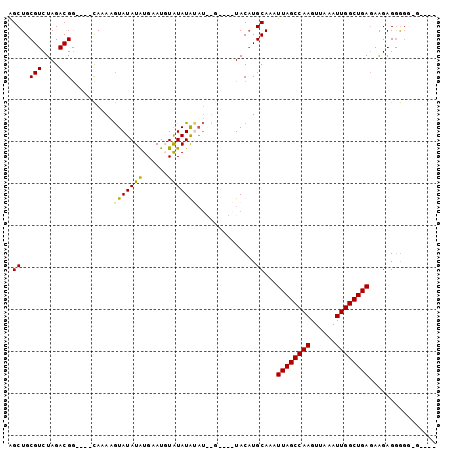
>X_DroMel_CAF1 15114188 97 - 22224390 AGCUGCGUCUAGACGG----CAAAAGUAUAUAUGAAUUUAUAUACAUAUAU-UGUACAUGCAAAUUAGCCAAGUUAAAUUGGCUGAGAGGAGGGGCGGAGGC ..((.(((((...(.(----((.....(((((((.((....)).)))))))-......)))...((((((((......))))))))..)...))))).)).. ( -26.90) >DroSec_CAF1 25914 86 - 1 AGCUGCGUCUAGACGAAGUAUACAUAUAUAUAUGAAUUUAUAUA----------UACAUGCAAAUUAGCCAAGUUAAAUUGGCUGAGAAGAGGAGA------ ..((.(.(((.......((((...((((((((......))))))----------)).))))...((((((((......))))))))..))).))).------ ( -20.40) >DroEre_CAF1 33080 88 - 1 GGCUGCGUCUAGACGG----CAAAAGUAUAUGUGCAUGUGUAUGUAU--G----UACAUGCAAAUUAGCCAAGUUAAAUUGGCUGAGAAGAGAGUG-GC--- .((..(.(((......----........((((((((((......)))--)----))))))....((((((((......))))))))....))))..-))--- ( -26.40) >DroYak_CAF1 32993 97 - 1 AGCUGCGUCUAGACGG----CAAAAGUAUAUAUGAAUGUAUAUGUAUAUGUACAUACUUGCAAAUUAGCCAAGUUAAAUUGGCUGAUAAGAGGGGG-GGGGC .(((.(.(((.....(----(((..(((((((((.((....)).)))))))))....))))..(((((((((......))))))))).....))).-).))) ( -27.50) >consensus AGCUGCGUCUAGACGG____CAAAAGUAUAUAUGAAUGUAUAUAUAU__G____UACAUGCAAAUUAGCCAAGUUAAAUUGGCUGAGAAGAGGGGG_G____ .((..(((....)))..........(((((((......)))))))..............))...((((((((......))))))))................ (-15.88 = -15.38 + -0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:55 2006