| Sequence ID | X_DroMel_CAF1 |
|---|---|
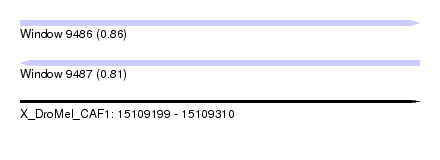
| Location | 15,109,199 – 15,109,310 |
| Length | 111 |
| Max. P | 0.864307 |

| Location | 15,109,199 – 15,109,310 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.56 |
| Mean single sequence MFE | -29.15 |
| Consensus MFE | -16.83 |
| Energy contribution | -17.07 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
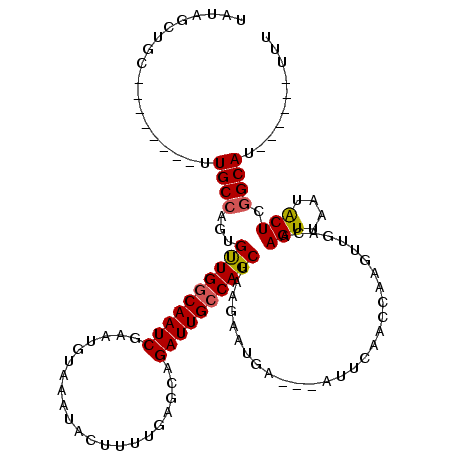
>X_DroMel_CAF1 15109199 111 + 22224390 AAA------AUGCCGAGCAUUAACUUGUCAACUUGGUUGAAU---UCAUUCUUAGCUGGCAAUCUGCUCAAAAGUAUUUACAUUCGAUUGCCAACAGUGGCAUCGUCAUAUGCAGCUAUA ...------..((.(((((.....((((((.((.((.((...---.))..)).)).))))))..)))))....((((.......((((.((((....))))))))....)))).)).... ( -25.80) >DroSec_CAF1 21038 117 + 1 AAACCGUUAAUGCCGAGCAUUAACUUGUCAACUUGGUUGAAU---UCAUUCUUAGCUGCCAAUCUGCUCAAAAGUAUUUACAUUCGAUUGCCAACACUGGCACAGUCAUAUGCAGCUAUU .....((((((((...))))))))...(((((...)))))..---.......(((((((.....((((....)))).........((((((((....)))))..)))....))))))).. ( -29.20) >DroEre_CAF1 28706 105 + 1 AAA------GUGCGGAGUAUUAACUUGUAAGCUGGCUUGAAUGGCUCAUUGCUGGCUGGCAAUCUGCGCAGCAGUGUUUACACUCGAUUGCCAACUCUGGCAA---------CAGUUAUA ...------(((((((((....))((((.(((..((.(((.....)))..))..))).)))))))))))(((((((....)))).(.((((((....))))))---------).)))... ( -34.70) >DroYak_CAF1 27204 103 + 1 AAA------AUGCCGAGUAUUAACUUGUCAACUUGGUUCAAU---UCAUUCUUAGCUGGCAAUCUGCUCGCAAGUAUUUACAUUCGAUUGCCAGCACUGGCAA--------GCAGCUAUA ...------..(((((((............))))))).....---.........((((((((((((....)).((....))....))))))))))..((((..--------...)))).. ( -26.90) >consensus AAA______AUGCCGAGCAUUAACUUGUCAACUUGGUUGAAU___UCAUUCUUAGCUGGCAAUCUGCUCAAAAGUAUUUACAUUCGAUUGCCAACACUGGCAA________GCAGCUAUA ..........(((.(((((.....((((((.((.((.(((.....)))..)).)).))))))..)))))...................(((((....))))).........)))...... (-16.83 = -17.07 + 0.25)



| Location | 15,109,199 – 15,109,310 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.56 |
| Mean single sequence MFE | -30.00 |
| Consensus MFE | -17.91 |
| Energy contribution | -17.97 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15109199 111 - 22224390 UAUAGCUGCAUAUGACGAUGCCACUGUUGGCAAUCGAAUGUAAAUACUUUUGAGCAGAUUGCCAGCUAAGAAUGA---AUUCAACCAAGUUGACAAGUUAAUGCUCGGCAU------UUU ....(((((((.((((.........((((((((((.....((((....))))....)))))))))).........---..(((((...)))))...))))))))..)))..------... ( -28.20) >DroSec_CAF1 21038 117 - 1 AAUAGCUGCAUAUGACUGUGCCAGUGUUGGCAAUCGAAUGUAAAUACUUUUGAGCAGAUUGGCAGCUAAGAAUGA---AUUCAACCAAGUUGACAAGUUAAUGCUCGGCAUUAACGGUUU ..(((((((......(((((((......)))..(((((.((....)).)))))))))....))))))).......---..(((((...)))))...((((((((...))))))))..... ( -32.80) >DroEre_CAF1 28706 105 - 1 UAUAACUG---------UUGCCAGAGUUGGCAAUCGAGUGUAAACACUGCUGCGCAGAUUGCCAGCCAGCAAUGAGCCAUUCAAGCCAGCUUACAAGUUAAUACUCCGCAC------UUU ..((((((---------((((..(.((((((((((..(((((........))))).))))))))))).)))))((((...........))))...)))))...........------... ( -29.90) >DroYak_CAF1 27204 103 - 1 UAUAGCUGC--------UUGCCAGUGCUGGCAAUCGAAUGUAAAUACUUGCGAGCAGAUUGCCAGCUAAGAAUGA---AUUGAACCAAGUUGACAAGUUAAUACUCGGCAU------UUU .........--------.((((((((((((((((((..(((((....)))))..).)))))))))).........---..........(((((....)))))))).)))).------... ( -29.10) >consensus UAUAGCUGC________UUGCCAGUGUUGGCAAUCGAAUGUAAAUACUUUUGAGCAGAUUGCCAGCUAAGAAUGA___AUUCAACCAAGUUGACAAGUUAAUACUCGGCAU______UUU ..................((((...((((((((((.....................)))))))))).............................(((....))).)))).......... (-17.91 = -17.97 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:47 2006