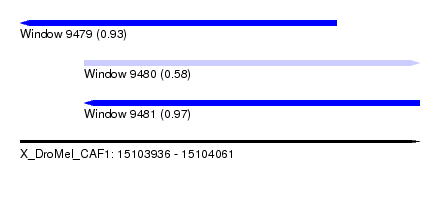
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,103,936 – 15,104,061 |
| Length | 125 |
| Max. P | 0.972569 |

| Location | 15,103,936 – 15,104,035 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.57 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -27.05 |
| Energy contribution | -27.17 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15103936 99 - 22224390 GGCAGGCGCUCGCGCAUUUAUUUCGCUUGUAAACGAGCGCAAUUUGCAAUUUGAGCGCGUCCGCCUCGCGUGUACCUUC------------------CCUA-CUUU--CUCCCUUGCUUU ((.(((..(.((((((...(((.((((((....)))))).))).))).....(((.((....)))))))).)..))).)------------------)...-....--............ ( -31.90) >DroSec_CAF1 16700 99 - 1 GGCAGGCGCUCGCGCAUUUAUUUCGCUUGUAAACGAGCGCAAUUUGCAAUUUGAGCGCGUCCGCCUCGCGUGUACCUUC------------------UUCA-CUUU--CUCCUUUGCUUU .(((((((..(((((.........(((((....)))))((.....)).......)))))..))))).))(((.......------------------..))-)...--............ ( -32.30) >DroSim_CAF1 20984 99 - 1 GGCAGGCGCUCGCGCAUUUAUUUCGCUUGUAAACGAGCGCAAUUUGCAAUUUGAGCGCGUCCGCCUCGCGUGUACCUUU------------------CCCA-CUUU--CUCCUUUGCUUU .(((((((..(((((.........(((((....)))))((.....)).......)))))..))))).))(((.......------------------..))-)...--............ ( -31.80) >DroEre_CAF1 24347 98 - 1 GGCGGGCGCUCGCGCAUUUAUUUCGCUUGUAAACGAGCGCAAUUUGCAAUUUGAGCGCGUCCGCCUCGCGUGUACCUC-------------------CCCA-CUUU--CUCCUUUGCUUU .(((((((..(((((.........(((((....)))))((.....)).......)))))..)))).)))(((......-------------------..))-)...--............ ( -34.10) >DroYak_CAF1 21811 117 - 1 GGCAGGCGCUCGCGCAUUUAUUUCGCUUGUAAACGAGCGCAAUUUGCAAUUUGAGCGCGUUCGCCUCGCGUGUACCUUCCACUUCCACUUCCACUUUCCUA-CUUU--CUCCUUUGCUUU ((.(((..(.((((((...(((.((((((....)))))).))).))).....(((.((....)))))))).)..))).)).....................-....--............ ( -32.80) >DroPer_CAF1 58517 101 - 1 GGCAGCUCCGCGCGUAUUUGUCUCGCUUGUAAAGGAGCGCAAUUUUCAAUUUAAGCGCGCUCGUUUCGCGUGUGCCUC-------------------UGCAUCCUUGGCACCGAUGCCUU ((((....((((((.........)))..(.(((.((((((..((........))..)))))).))))))).(((((..-------------------.........)))))...)))).. ( -31.50) >consensus GGCAGGCGCUCGCGCAUUUAUUUCGCUUGUAAACGAGCGCAAUUUGCAAUUUGAGCGCGUCCGCCUCGCGUGUACCUUC__________________CCCA_CUUU__CUCCUUUGCUUU .(((((((..(((((.........(((((....)))))((.....)).......)))))..))))).))................................................... (-27.05 = -27.17 + 0.11)

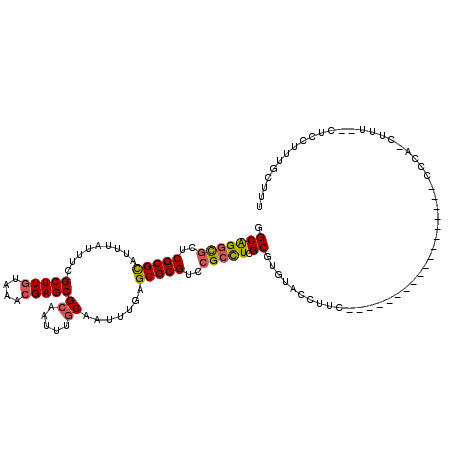

| Location | 15,103,956 – 15,104,061 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.45 |
| Mean single sequence MFE | -34.97 |
| Consensus MFE | -18.54 |
| Energy contribution | -18.87 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15103956 105 + 22224390 -GAAGGUACACGCGAGGCGGACGCGCUCAAAUUGCAAAUUGCGCUCGUUUACAAGCGAAAUAAAUGCGCGAGCGCCUGCCAUUUGGCAAAAGUCAGCCA--------------UGCUCCA -...((.....(((.(((.(((((((((.....((.....))((.((((((.........)))))).)))))))).((((....))))...))).))).--------------))).)). ( -37.70) >DroPse_CAF1 31419 109 + 1 -----------GCGAAACGAACGCGCUUAAAUUGCAAAUUGCGCUCCUUUACAAGCGAGACAAAUACGCGCGGACCUGCCAUUUGGCGAAAGUCGGCCAUCCCACGCCAGACCUACGCCA -----------(((.......(((((.......((.....))((((((.....)).))).)......)))))(((.((((....))))...)))(((........))).......))).. ( -28.40) >DroSim_CAF1 21004 105 + 1 -AAAGGUACACGCGAGGCGGACGCGCUCAAAUUGCAAAUUGCGCUCGUUUACAAGCGAAAUAAAUGCGCGAGCGCCUGCCAUUUGGCAAAAGUCAGCCA--------------UGCUCCA -...((.....(((.(((.(((((((((.....((.....))((.((((((.........)))))).)))))))).((((....))))...))).))).--------------))).)). ( -37.70) >DroEre_CAF1 24367 104 + 1 --GAGGUACACGCGAGGCGGACGCGCUCAAAUUGCAAAUUGCGCUCGUUUACAAGCGAAAUAAAUGCGCGAGCGCCCGCCAUUUGGCAGGAGUCAGCCA--------------UGCCCCA --..((.....(((.(((.(((((((((.....((.....))((.((((((.........)))))).))))))))(((((....))).)).))).))).--------------))).)). ( -37.80) >DroYak_CAF1 21848 106 + 1 GGAAGGUACACGCGAGGCGAACGCGCUCAAAUUGCAAAUUGCGCUCGUUUACAAGCGAAAUAAAUGCGCGAGCGCCUGCCAUUUGGCAAGAGUCAGGCA--------------UGCUCCA ....((..((.((.(((((..(((((.......((.....))..(((((....))))).......)))))..)))))(((....))).........)).--------------))..)). ( -35.30) >DroPer_CAF1 58540 108 + 1 --GAGGCACACGCGAAACGAGCGCGCUUAAAUUGAAAAUUGCGCUCCUUUACAAGCGAGACAAAUACGCGCGGAGCUGCCAUUUG----------GCCAUCCCACGCCAGACCUACGCCA --..(((...(((.(((.(((((((.((........)).))))))).)))....)))..........(((.(((...(((....)----------))..)))..))).........))). ( -32.90) >consensus __AAGGUACACGCGAGGCGAACGCGCUCAAAUUGCAAAUUGCGCUCGUUUACAAGCGAAAUAAAUGCGCGAGCGCCUGCCAUUUGGCAAAAGUCAGCCA______________UGCUCCA ....(((...(((.((((((..((((..............))))))))))....)))...((((((.(((......)))))))))..........)))...................... (-18.54 = -18.87 + 0.34)

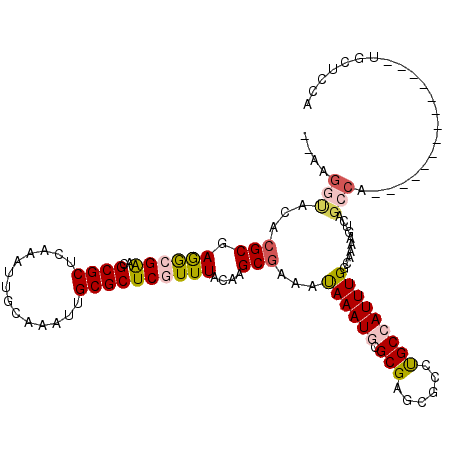

| Location | 15,103,956 – 15,104,061 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.45 |
| Mean single sequence MFE | -41.28 |
| Consensus MFE | -25.78 |
| Energy contribution | -26.37 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972569 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15103956 105 - 22224390 UGGAGCA--------------UGGCUGACUUUUGCCAAAUGGCAGGCGCUCGCGCAUUUAUUUCGCUUGUAAACGAGCGCAAUUUGCAAUUUGAGCGCGUCCGCCUCGCGUGUACCUUC- .((.(((--------------((((.(((..(((((....)))))(((((((.(((...(((.((((((....)))))).))).)))....)))))))))).)))....)))).))...- ( -42.60) >DroPse_CAF1 31419 109 - 1 UGGCGUAGGUCUGGCGUGGGAUGGCCGACUUUCGCCAAAUGGCAGGUCCGCGCGUAUUUGUCUCGCUUGUAAAGGAGCGCAAUUUGCAAUUUAAGCGCGUUCGUUUCGC----------- (((((.(((((.(((........)))))))).)))))...((.(.(..(((((....((((..(((((......)))))......)))).....)))))..).).))..----------- ( -36.60) >DroSim_CAF1 21004 105 - 1 UGGAGCA--------------UGGCUGACUUUUGCCAAAUGGCAGGCGCUCGCGCAUUUAUUUCGCUUGUAAACGAGCGCAAUUUGCAAUUUGAGCGCGUCCGCCUCGCGUGUACCUUU- .((.(((--------------((((.(((..(((((....)))))(((((((.(((...(((.((((((....)))))).))).)))....)))))))))).)))....)))).))...- ( -42.60) >DroEre_CAF1 24367 104 - 1 UGGGGCA--------------UGGCUGACUCCUGCCAAAUGGCGGGCGCUCGCGCAUUUAUUUCGCUUGUAAACGAGCGCAAUUUGCAAUUUGAGCGCGUCCGCCUCGCGUGUACCUC-- .((..((--------------((((.(((..(((((....)))))(((((((.(((...(((.((((((....)))))).))).)))....)))))))))).)))....)))..))..-- ( -45.00) >DroYak_CAF1 21848 106 - 1 UGGAGCA--------------UGCCUGACUCUUGCCAAAUGGCAGGCGCUCGCGCAUUUAUUUCGCUUGUAAACGAGCGCAAUUUGCAAUUUGAGCGCGUUCGCCUCGCGUGUACCUUCC .((.(((--------------(((........((((....))))((((..(((((.........(((((....)))))((.....)).......)))))..))))..)))))).)).... ( -43.40) >DroPer_CAF1 58540 108 - 1 UGGCGUAGGUCUGGCGUGGGAUGGC----------CAAAUGGCAGCUCCGCGCGUAUUUGUCUCGCUUGUAAAGGAGCGCAAUUUUCAAUUUAAGCGCGCUCGUUUCGCGUGUGCCUC-- .((((........(((((((...((----------(....)))...)))))))..........(((.((.(((.((((((..((........))..)))))).))))).)))))))..-- ( -37.50) >consensus UGGAGCA______________UGGCUGACUUUUGCCAAAUGGCAGGCGCUCGCGCAUUUAUUUCGCUUGUAAACGAGCGCAAUUUGCAAUUUGAGCGCGUCCGCCUCGCGUGUACCUU__ .....................((((........))))....(((((((..(((((.........(((((....)))))((.....)).......)))))..))))).))........... (-25.78 = -26.37 + 0.59)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:42 2006