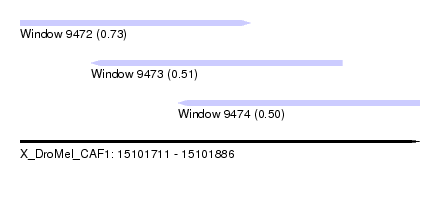
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,101,711 – 15,101,886 |
| Length | 175 |
| Max. P | 0.733616 |

| Location | 15,101,711 – 15,101,812 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
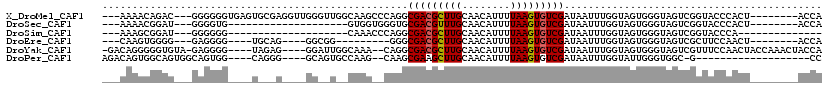
| Mean pairwise identity | 79.18 |
| Mean single sequence MFE | -36.35 |
| Consensus MFE | -11.44 |
| Energy contribution | -12.11 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.31 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15101711 101 + 22224390 GGAAUACGGCCGUCUUUGU--------CUGCAU-UAAUGCAUAUGCAUGA--GACUUAAGCGACGGCCACGCCUCGAAGUUGGU--------AGUGGGUACCGACUACCCACUACCAAAU ((.....(((((((...((--------((((((-........))))...)--)))......)))))))...))......(((((--------((((((((.....))))))))))))).. ( -42.30) >DroSec_CAF1 14527 102 + 1 GGAAUACGGCCGUCUUUGU--------CUGCAU-UAAUGCAUAUGCAUGAGAGACUUAAGCG-UGGCCACGCCUCGCAGUUGGU--------AGUGGGUACCGACUACCCACUACCAAAU ((.......))((((((..--------.(((((-........)))))..))))))....(((-.(((...))).)))..(((((--------((((((((.....))))))))))))).. ( -41.10) >DroSim_CAF1 18819 95 + 1 GGAAUACGGCCGUCUUUGU--------CUGCAU-UAAUGCAUAUGCAUGAGAGACUUAAGCG-UGGCCACGCCUCGCAG---------------UGGGUACCGACUACCCACUACCAAAU ((.........((((((..--------.(((((-........)))))..))))))....(((-.(((...))).)))((---------------((((((.....)))))))).)).... ( -32.50) >DroEre_CAF1 22274 100 + 1 GGAAUACGGCCGUCUUUGU--------CUGCAU-UAAUGCAUAUGCAUGA--GACUUAAGCG-CGGCCACGCCUCGAAGUUGGU--------AGUUGGAAGCGACUACCCACUACCAAAU ((.....(((((((((.((--------((((((-........))))...)--)))..))).)-))))).........(((.(((--------(((((....)))))))).))).)).... ( -37.20) >DroYak_CAF1 19641 116 + 1 GGAAUACGGCCUUCUUUGUCUGCUUGUCUGCAU-UAAUGCAUAUGCAUGA--GACUUAAGCG-CGGCCACGCCUCGAAGUUGGUAGUUUGGUAGUUGGAAACGACUACCCACUACCAAAU ((.....((((..(((.((((((......))..-..((((....)))).)--)))..)))..-.))))...))......((((((((..((((((((....)))))))).)))))))).. ( -43.90) >DroPer_CAF1 55746 86 + 1 -GCAUCCCAC--UCUUCUU--------CUGUGUGUUAUGCAUAUGCAUGA--GUCUUAAGCA-GGGCCACACCUCGAGGUGG-------------------C-GCCACCCAAUACCAAAU -((....(((--.(.....--------..).)))((((((....))))))--.......)).-(((((((........))))-------------------)-.)).............. ( -21.10) >consensus GGAAUACGGCCGUCUUUGU________CUGCAU_UAAUGCAUAUGCAUGA__GACUUAAGCG_CGGCCACGCCUCGAAGUUGGU________AGUGGGUACCGACUACCCACUACCAAAU ((.....((((.................(((((...)))))...((.(((.....))).))...)))).........(((((...................)))))........)).... (-11.44 = -12.11 + 0.67)

| Location | 15,101,742 – 15,101,852 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.30 |
| Mean single sequence MFE | -37.75 |
| Consensus MFE | -15.73 |
| Energy contribution | -16.29 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.42 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15101742 110 - 22224390 GCAAGCCCAGGCGACGCUUGCAACAUUUUAAGUGUCGAUAAUUUGGUAGUGGGUAGUCGGUACCCACU--------ACCAACUUCGAGGCGUGGCCGUCGCUUAAGUC--UCAUGCAUAU (((.((....))((((((((........))))))))((....(((((((((((((.....))))))))--------)))))..))(((((..(((....)))...)))--)).))).... ( -45.80) >DroSec_CAF1 14558 110 - 1 -GUGGUGGGUGCGACGUUUGCAACAUUUUAAGUGUCGAUAAUUUGGUAGUGGGUAGUCGGUACCCACU--------ACCAACUGCGAGGCGUGGCCA-CGCUUAAGUCUCUCAUGCAUAU -((((((((...(((...((((((((.....)))).......(((((((((((((.....))))))))--------))))).))))((((((....)-)))))..))).))))).))).. ( -40.20) >DroSim_CAF1 18850 103 - 1 -CAAACCCAGGCGACGCUUGCAACAUUUUAAGUGUCGAUAAUUUGGUAGUGGGUAGUCGGUACCCA---------------CUGCGAGGCGUGGCCA-CGCUUAAGUCUCUCAUGCAUAU -((((......(((((((((........)))))))))....))))((((((((((.....))))))---------------))))(((((..(((..-.)))...))))).......... ( -36.50) >DroEre_CAF1 22305 101 - 1 --------GGGCGACGCUUGCAACAUUUUAAGUGUCGAUAAUUUGGUAGUGGGUAGUCGCUUCCAACU--------ACCAACUUCGAGGCGUGGCCG-CGCUUAAGUC--UCAUGCAUAU --------..((((((((((........))))))))((....(((((((((((........))).)))--------)))))..))(((((..(((..-.)))...)))--))..)).... ( -33.40) >DroYak_CAF1 19680 115 - 1 GCAAA--CAGGCGACGCUUGCAACAUUUUAAGUGUCGAUAAUUUGGUAGUGGGUAGUCGUUUCCAACUACCAAACUACCAACUUCGAGGCGUGGCCG-CGCUUAAGUC--UCAUGCAUAU (((..--.((((((((((((........))))))))((....((((((((.((((((........))))))..))))))))..)).((((((....)-)))))..)))--)..))).... ( -40.80) >DroPer_CAF1 55775 95 - 1 CCAAG--CAAGCGAAGCUUGCAACAUUUUAAGUGUCGAUAAUUUGGUAUUGGGUGGC-G-------------------CCACCUCGAGGUGUGGCCC-UGCUUAAGAC--UCAUGCAUAU ....(--(((((...))))))....((((((((.((((....))))....((((.((-(-------------------((.......))))).))))-.)))))))).--.......... ( -29.80) >consensus _CAAG__CAGGCGACGCUUGCAACAUUUUAAGUGUCGAUAAUUUGGUAGUGGGUAGUCGGUACCCACU________ACCAACUUCGAGGCGUGGCCG_CGCUUAAGUC__UCAUGCAUAU .........(((.((((((.(.((((.....))))((((.((((......)))).))))..........................))))))).)))...((.............)).... (-15.73 = -16.29 + 0.56)


| Location | 15,101,780 – 15,101,886 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 64.01 |
| Mean single sequence MFE | -30.58 |
| Consensus MFE | -9.29 |
| Energy contribution | -9.32 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.30 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15101780 106 - 22224390 ---AAAACAGAC---GGGGGGUGAGUGCGAGGUUGGGUUGGCAAGCCCAGGCGACGCUUGCAACAUUUUAAGUGUCGAUAAUUUGGUAGUGGGUAGUCGGUACCCACU--------ACCA ---......(((---(.((((((..((((((..((((((....))))))(....).)))))).))))))...)))).......((((((((((((.....))))))))--------)))) ( -42.70) >DroSec_CAF1 14597 86 - 1 ---AAAACGGAU---GGGGUG--------------------GUGGUGGGUGCGACGUUUGCAACAUUUUAAGUGUCGAUAAUUUGGUAGUGGGUAGUCGGUACCCACU--------ACCA ---.........---....((--------------------((((((((((((((...(((..((((((((((.......)))))).)))).)))))).)))))))))--------)))) ( -29.80) >DroSim_CAF1 18888 80 - 1 ---AAAGCGGAU---GGGGGG--------------------CAAACCCAGGCGACGCUUGCAACAUUUUAAGUGUCGAUAAUUUGGUAGUGGGUAGUCGGUACCCA-------------- ---....(((((---.(((..--------------------....)))...(((((((((........)))))))))...)))))....((((((.....))))))-------------- ( -24.30) >DroEre_CAF1 22342 89 - 1 ---CAAGUGGGG---GAGGGG----UGCAG----GGCGG---------GGGCGACGCUUGCAACAUUUUAAGUGUCGAUAAUUUGGUAGUGGGUAGUCGCUUCCAACU--------ACCA ---..(((..((---(((((.----(((..----.((.(---------((.(((((((((........)))))))))....))).)).....))).)).))))).)))--------.... ( -27.10) >DroYak_CAF1 19717 108 - 1 -GACAGGGGGUGUA-GAGGGG----UAGAG----GGAUUGGCAAA--CAGGCGACGCUUGCAACAUUUUAAGUGUCGAUAAUUUGGUAGUGGGUAGUCGUUUCCAACUACCAAACUACCA -.............-....((----(((..----...........--....(((((((((........)))))))))....((((((((((((........))).)))))))))))))). ( -32.30) >DroPer_CAF1 55812 90 - 1 AGACAGUGGCAGUGGCAGUGG----CAGGG----GCAGUGCCAAG--CAAGCGAAGCUUGCAACAUUUUAAGUGUCGAUAAUUUGGUAUUGGGUGGC-G-------------------CC .......(((..(((((.((.----(...)----.)).))))).(--(((((...))))))..((((...(((..(((....)))..))).))))..-)-------------------)) ( -27.30) >consensus ___AAAGCGGAG___GAGGGG____U___G____GG__U__CAAG__CAGGCGACGCUUGCAACAUUUUAAGUGUCGAUAAUUUGGUAGUGGGUAGUCGGUACCCACU________ACCA ...................................................(((((((((........)))))))))........................................... ( -9.29 = -9.32 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:35 2006