
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,082,464 – 15,082,583 |
| Length | 119 |
| Max. P | 0.893851 |

| Location | 15,082,464 – 15,082,583 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.74 |
| Mean single sequence MFE | -35.40 |
| Consensus MFE | -24.25 |
| Energy contribution | -25.26 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.678605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
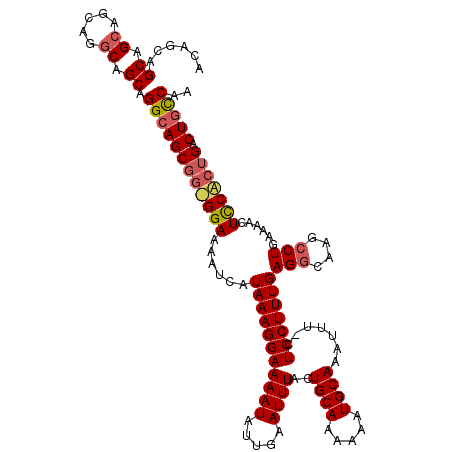
>X_DroMel_CAF1 15082464 119 + 22224390 UUGGCAGUCAGCGGAGUUUUCAGGCUUGCCUCAAAGGAG-AAAUUUGCAUUUUUUGCAGUAAAUUCAAUAUUUUCCUUUAUGAUUUUCCGCCGCUGCCUGCUGCCUGCUGCUGCUGCUGU ..(((((.((((((.((...(((((..((...(((((((-((...((((.....))))(......)....)))))))))..(......)...)).)))))..)))))))))))))..... ( -38.40) >DroAna_CAF1 35047 114 + 1 GGGAAAGUCAGAGGAGA-UUCAGGCUCGCCUCAAAGGAG-AAAUUUGCAUUUUUUGCAGUAAAUUCAAUAUUUUCCUUUAUGAUUUUCC---GCUGCCUGCUGU-UGCUGCUGCUGCUGA .(((((((((((((.((-.......)).))))(((((((-((...((((.....))))(......)....))))))))).)))))))))---((.((..((...-.)).)).))...... ( -35.20) >DroYak_CAF1 36476 117 + 1 UUGGCAGUCAGUGAAGUUUUCAGGCUUGGCUCAAAGGAAAAAAUUUGCAUUUUUUGCAGUAAAUUCAAUAUUUUCCUUUAUGAUUUUCCACCGCUGCCUGCUGCC---UGCUGCUGCUGU ..(((((.((((...((...(((((..(((..(((((((((.(((((((.....))))........))).))))))))).((......))..))))))))..)).---.))))))))).. ( -32.60) >consensus UUGGCAGUCAGAGGAGUUUUCAGGCUUGCCUCAAAGGAG_AAAUUUGCAUUUUUUGCAGUAAAUUCAAUAUUUUCCUUUAUGAUUUUCC_CCGCUGCCUGCUGCCUGCUGCUGCUGCUGU ..(((((.((((((......(((((.......(((((((.....(((((.....))))).............)))))))..(.....).......))))).......))))))))))).. (-24.25 = -25.26 + 1.00)



| Location | 15,082,464 – 15,082,583 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.74 |
| Mean single sequence MFE | -31.73 |
| Consensus MFE | -25.29 |
| Energy contribution | -26.63 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15082464 119 - 22224390 ACAGCAGCAGCAGCAGGCAGCAGGCAGCGGCGGAAAAUCAUAAAGGAAAAUAUUGAAUUUACUGCAAAAAAUGCAAAUUU-CUCCUUUGAGGCAAGCCUGAAAACUCCGCUGACUGCCAA ......((.((.....)).)).((((((((((((......(((((((((((.(((.((((........)))).)))))))-.)))))))(((....)))......))))))).))))).. ( -37.40) >DroAna_CAF1 35047 114 - 1 UCAGCAGCAGCAGCA-ACAGCAGGCAGC---GGAAAAUCAUAAAGGAAAAUAUUGAAUUUACUGCAAAAAAUGCAAAUUU-CUCCUUUGAGGCGAGCCUGAA-UCUCCUCUGACUUUCCC ......((.((.((.-...))..)).))---(((((.(((.((((((((((.(((.((((........)))).)))))))-.))))))((((.((.......-)).))))))).))))). ( -25.70) >DroYak_CAF1 36476 117 - 1 ACAGCAGCAGCA---GGCAGCAGGCAGCGGUGGAAAAUCAUAAAGGAAAAUAUUGAAUUUACUGCAAAAAAUGCAAAUUUUUUCCUUUGAGCCAAGCCUGAAAACUUCACUGACUGCCAA ...((.((....---.)).)).((((((((((((...(((((((((((((.(((........((((.....))))))).)))))))))).((...)).)))....))))))).))))).. ( -32.10) >consensus ACAGCAGCAGCAGCAGGCAGCAGGCAGCGG_GGAAAAUCAUAAAGGAAAAUAUUGAAUUUACUGCAAAAAAUGCAAAUUU_CUCCUUUGAGGCAAGCCUGAAAACUCCACUGACUGCCAA ......((.((.....)).)).((((((((((((......(((((((((((.....))))..((((.....)))).......)))))))(((....)))......))))))).))))).. (-25.29 = -26.63 + 1.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:26 2006