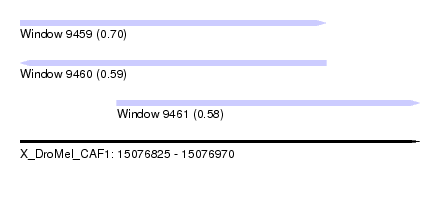
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,076,825 – 15,076,970 |
| Length | 145 |
| Max. P | 0.699429 |

| Location | 15,076,825 – 15,076,936 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 81.06 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -21.85 |
| Energy contribution | -20.97 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
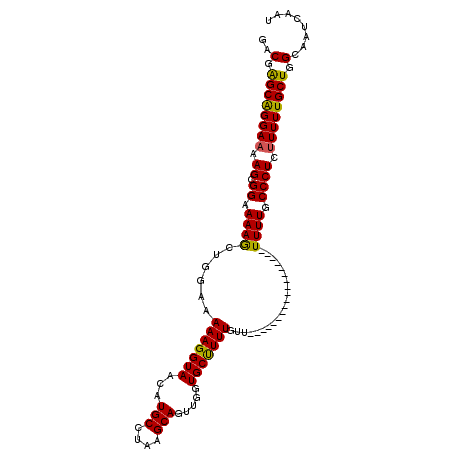
>X_DroMel_CAF1 15076825 111 + 22224390 UUCGGGGAAUCAGUUGGAAGAGGCAUGGUUAGCGGGACGAGCAGGAAAAGCGGAAAAGCUGGAAAAAGGUAACAUGCCUAAGCAGUUUGUGCUUUUGUUGUUUUUUUUUUU (((.(..((((((((......))).)))))..).))).((((((.(((((((...((((((.....((((.....))))...)))))).))))))).))))))........ ( -27.10) >DroSim_CAF1 24021 100 + 1 UUCGGGGAACCAGUUGGGGUAGGCAUGGUUAGCGGGACGAGCGGGAAAAGCGGAAAAGCUGGAAAAAGGUAACAUGCCUAAGCAGUUGGUGCUUUUGUUU----------- ...((((.((((..((...((((((((....((.......))......(((......)))............))))))))..))..)))).)))).....----------- ( -26.80) >DroEre_CAF1 28999 98 + 1 UUCGGGGAAGCAGCUGGGAAAGGUAUGGGAAGCGGUACCGGCAGGAAAAGCGGAAAAACUGGAAAAAGGUAACAUGCCUAAGCAGUUGUUGCCUUUGU------------- (((((.......(((..(...(((((.(....).)))))..)......))).......))))).(((((((((((((....)))..))))))))))..------------- ( -28.54) >consensus UUCGGGGAACCAGUUGGGAAAGGCAUGGUUAGCGGGACGAGCAGGAAAAGCGGAAAAGCUGGAAAAAGGUAACAUGCCUAAGCAGUUGGUGCUUUUGUU____________ ...((((.((((((((....(((((((....((.......))........(((.....)))...........)))))))...)))))))).))))................ (-21.85 = -20.97 + -0.88)



| Location | 15,076,825 – 15,076,936 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 81.06 |
| Mean single sequence MFE | -15.93 |
| Consensus MFE | -12.97 |
| Energy contribution | -12.87 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15076825 111 - 22224390 AAAAAAAAAAACAACAAAAGCACAAACUGCUUAGGCAUGUUACCUUUUUCCAGCUUUUCCGCUUUUCCUGCUCGUCCCGCUAACCAUGCCUCUUCCAACUGAUUCCCCGAA .................(((((.....)))))(((((((............(((......)))......((.......))....))))))).................... ( -15.30) >DroSim_CAF1 24021 100 - 1 -----------AAACAAAAGCACCAACUGCUUAGGCAUGUUACCUUUUUCCAGCUUUUCCGCUUUUCCCGCUCGUCCCGCUAACCAUGCCUACCCCAACUGGUUCCCCGAA -----------..........((((..((..((((((((............(((......)))......((.......))....))))))))...))..))))........ ( -17.60) >DroEre_CAF1 28999 98 - 1 -------------ACAAAGGCAACAACUGCUUAGGCAUGUUACCUUUUUCCAGUUUUUCCGCUUUUCCUGCCGGUACCGCUUCCCAUACCUUUCCCAGCUGCUUCCCCGAA -------------..(((((.((((..(((....))))))).)))))...(((((.....((.......)).((((..........))))......))))).......... ( -14.90) >consensus ____________AACAAAAGCACCAACUGCUUAGGCAUGUUACCUUUUUCCAGCUUUUCCGCUUUUCCUGCUCGUCCCGCUAACCAUGCCUAUCCCAACUGAUUCCCCGAA .................(((((.....)))))(((((((............(((......)))......((.......))....))))))).................... (-12.97 = -12.87 + -0.11)

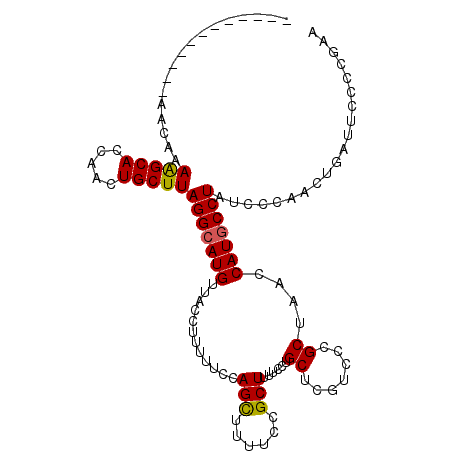

| Location | 15,076,860 – 15,076,970 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.79 |
| Mean single sequence MFE | -28.07 |
| Consensus MFE | -17.21 |
| Energy contribution | -16.66 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15076860 110 + 22224390 GACGAGCAGGAAAAGCGGAAAAGCUGGAAAAAGGUAACAUGCCUAAGCAGUUUGUGCUUUUGUUGUUUUUUUUUUUUUUUUGUUUUGCCCUCCUUUUGCUGGCAAUCAAU (.(.(((((((..((.((.(((((..((((((((.((((.((..(((((.....)))))..))))))....))))))))..))))).))))..))))))).))....... ( -27.70) >DroSim_CAF1 24056 93 + 1 GACGAGCGGGAAAAGCGGAAAAGCUGGAAAAAGGUAACAUGCCUAAGCAGUUGGUGCUUUUGUUU-----------------UUUUGCCCUCUUUUUGCUGGCAAUCAAU (.(.((((((((..((((((((((.......((((.....))))(((((.....)))))..))))-----------------))))))....)))))))).))....... ( -23.90) >DroEre_CAF1 29034 91 + 1 UACCGGCAGGAAAAGCGGAAAAACUGGAAAAAGGUAACAUGCCUAAGCAGUUGUUGCCUUUGU-------------------UUUUGCCCUCUUUUUGCUGGCAAUCAAU ..((((((((((.((.((((((((.....(((((((((((((....)))..))))))))))))-------------------)))).)))).))))))))))........ ( -32.60) >consensus GACGAGCAGGAAAAGCGGAAAAGCUGGAAAAAGGUAACAUGCCUAAGCAGUUGGUGCUUUUGUU__________________UUUUGCCCUCUUUUUGCUGGCAAUCAAU ..(.((((((((.((.((.((((......(((((((...(((....))).....))))))).....................)))).)))).)))))))).)........ (-17.21 = -16.66 + -0.55)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:24 2006