
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,074,551 – 15,074,683 |
| Length | 132 |
| Max. P | 0.941461 |

| Location | 15,074,551 – 15,074,657 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.79 |
| Mean single sequence MFE | -20.67 |
| Consensus MFE | -11.80 |
| Energy contribution | -12.88 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
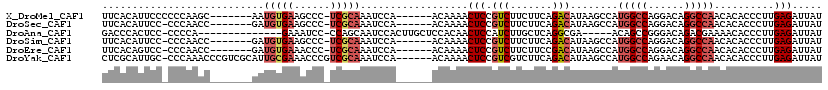
>X_DroMel_CAF1 15074551 106 - 22224390 UUCACAUUCCCCCCAAGC-------AAUGUGAAGCCC-UCGCAAAUCCA------ACAAAACUCCGUCUUCUUCAGACAUAAGCCAUGGCCAGGACAGGCCAACACACCCUUGAGAUUAU ((((((((.(......).-------))))))))((..-..)).((((((------(.........((((.....))))........(((((......)))))........))).)))).. ( -22.00) >DroSec_CAF1 35724 105 - 1 UUCACAUUCC-CCCAACC-------GAUGUGAAGCCC-UCGCAAAUCCA------ACAAAACUCCGUCUUCUUCAGACAUAAGCCAUGGCCAGGACAGGCCAACACACCCUUGAGAUUAU ((((((((..-.......-------))))))))((..-..)).((((((------(.........((((.....))))........(((((......)))))........))).)))).. ( -21.60) >DroAna_CAF1 28099 99 - 1 GACCCACUCC-CCCCA--------------GAAAUCC-CCAGCAAUCCACUUGCUCCACAACUCCAUCUUGCUCAGGCGA-----ACAGCCGGGACAGACGAAAACACCCUUGAGAUUAU ..........-.....--------------.......-..(((((.....)))))................(((((((..-----...)))(((.............))).))))..... ( -14.52) >DroSim_CAF1 21669 105 - 1 UUCACAUUCC-CCCAACC-------GAUGUGAAGCCC-UCGCAAAUCCA------ACAAAACUCCGUCUUCUUCAGACAUAAGCCAUGGCCAGGACAGGCCAACACACCCUUGAGAUUAU ((((((((..-.......-------))))))))((..-..)).((((((------(.........((((.....))))........(((((......)))))........))).)))).. ( -21.60) >DroEre_CAF1 26684 105 - 1 UUCACAGUCC-CCCAACC-------GAUGUGAAACCC-UCGCAAAUCCA------ACAAAACUCCGUCUUCUUCCGACAUAAGCCAUGGCCAGGACAGGCCAACACACCCUUGAGAUUAU ((((((.((.-.......-------))))))))...(-((.........------..........(((.......)))........(((((......)))))..........)))..... ( -18.90) >DroYak_CAF1 28605 113 - 1 CUCGCAUUGC-CCCAAACCCGUCGCAUUGCGAAACCCGUCGCAAAUCCA------ACAAAACUCCGUCGUCUUCAGACAUAAGCCAUGGCCAGAACAGGCCAACACACCCUUGAGAUUAU .(((((.(((-..(......)..))).)))))...........((((((------(.........(((.......)))........(((((......)))))........))).)))).. ( -25.40) >consensus UUCACAUUCC_CCCAACC_______GAUGUGAAACCC_UCGCAAAUCCA______ACAAAACUCCGUCUUCUUCAGACAUAAGCCAUGGCCAGGACAGGCCAACACACCCUUGAGAUUAU ...........................(((((......)))))..................(((.(((.......)))........(((((......)))))..........)))..... (-11.80 = -12.88 + 1.09)

| Location | 15,074,591 – 15,074,683 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -18.46 |
| Consensus MFE | -13.35 |
| Energy contribution | -13.79 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
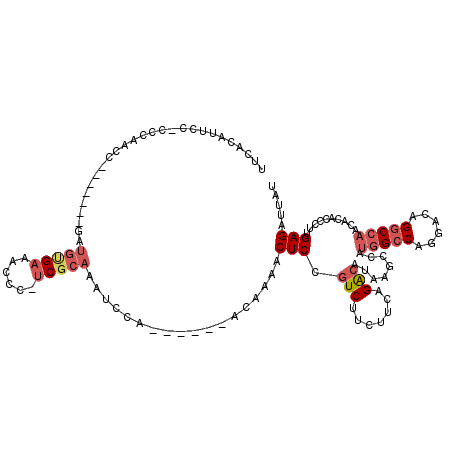
>X_DroMel_CAF1 15074591 92 - 22224390 UUACUCAUACGCCCCGUUGCUC-GGCAUUCACAUUCCCCCCAAGC-------AAUGUGAAGCCC-UCGCAAAUCCAACAAAACUCCGUCUUCUUCAGACAU ................((((..-(((.((((((((.(......).-------))))))))))).-..))))...............((((.....)))).. ( -19.00) >DroSec_CAF1 35764 91 - 1 UUACUCAUACGCCCCGUUGCUC-GGCAUUCACAUUCC-CCCAACC-------GAUGUGAAGCCC-UCGCAAAUCCAACAAAACUCCGUCUUCUUCAGACAU ................((((..-(((.((((((((..-.......-------))))))))))).-..))))...............((((.....)))).. ( -18.60) >DroSim_CAF1 21709 91 - 1 UUACUCAUACGCCCCGUUGCUC-GGCAUUCACAUUCC-CCCAACC-------GAUGUGAAGCCC-UCGCAAAUCCAACAAAACUCCGUCUUCUUCAGACAU ................((((..-(((.((((((((..-.......-------))))))))))).-..))))...............((((.....)))).. ( -18.60) >DroEre_CAF1 26724 91 - 1 UUACACAUACGCCCCGUUGCUC-GGCAUUCACAGUCC-CCCAACC-------GAUGUGAAACCC-UCGCAAAUCCAACAAAACUCCGUCUUCUUCCGACAU ................((((..-((..((((((.((.-.......-------))))))))..))-..))))...............(((.......))).. ( -13.10) >DroYak_CAF1 28645 100 - 1 UUACUCAUACGCCCCGUUGCUCCGGCACUCGCAUUGC-CCCAAACCCGUCGCAUUGCGAAACCCGUCGCAAAUCCAACAAAACUCCGUCGUCUUCAGACAU ................((((..(((...(((((.(((-..(......)..))).)))))...)))..))))...............(((.......))).. ( -23.00) >consensus UUACUCAUACGCCCCGUUGCUC_GGCAUUCACAUUCC_CCCAACC_______GAUGUGAAGCCC_UCGCAAAUCCAACAAAACUCCGUCUUCUUCAGACAU ................((((...(((.((((((((.................)))))))))))....))))...............(((.......))).. (-13.35 = -13.79 + 0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:18 2006