
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,070,971 – 15,071,069 |
| Length | 98 |
| Max. P | 0.808912 |

| Location | 15,070,971 – 15,071,069 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 87.38 |
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -25.15 |
| Energy contribution | -25.40 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
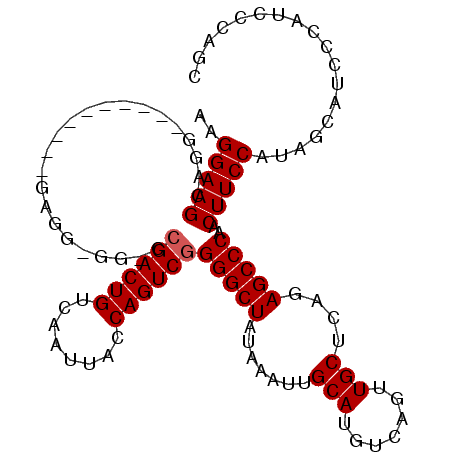
>X_DroMel_CAF1 15070971 98 + 22224390 AAGGAAGCAGG----------GAGGAAG-CAGACUGUCAAUUACCAGUCGGGGCUAUAAAUUGCAUGUCAGUUGCUCAGAGCCCAACUUCCAUAGCAUCCCAUCCCAGC ......((.((----------(((((.(-(.(((((........))))).(((((.......(((.......)))....)))))..........)).)))..)))).)) ( -31.10) >DroSec_CAF1 32044 97 + 1 AAGGAAGCAGG----------GAGG-GG-CCGACUGUCAAUUACCAGUCGGGGCUAUAAAUUGCAUGUCAGUUGCUCAGAGCCCAACUUCCAUAGCAUCCCAUCCCAAC ..(((.((..(----------((((-..-.((((((........))))))(((((.......(((.......)))....)))))..)))))...)).)))......... ( -33.10) >DroSim_CAF1 18359 97 + 1 AAGGAAGCGGG----------GAGG-GG-CCGACUGUCAAUUACCAGUCGGGGCUAUAAAUUGCAUGUCAGUUGCUCAGAGCCCAACUUCCAUAGCAUCCCAUCCCAGC ..(((.((..(----------((((-..-.((((((........))))))(((((.......(((.......)))....)))))..)))))...)).)))......... ( -34.20) >DroYak_CAF1 25451 102 + 1 AAGGAAGCAGGAGUGCGCGGGAAAG-GGA-CGACUGUCAAUUACCAGUCGGGGCUAUAAAUUGCAUGUCAGUUGCUCAGAGCCCAACUUCCAUGGCAUC-----CAAGC .........(((.(((.((((((..-...-((((((........))))))(((((.......(((.......)))....)))))...)))).)))))))-----).... ( -31.90) >consensus AAGGAAGCAGG__________GAGG_GG_CCGACUGUCAAUUACCAGUCGGGGCUAUAAAUUGCAUGUCAGUUGCUCAGAGCCCAACUUCCAUAGCAUCCCAUCCCAGC ..(((((.......................((((((........))))))(((((.......(((.......)))....)))))..))))).................. (-25.15 = -25.40 + 0.25)


| Location | 15,070,971 – 15,071,069 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 87.38 |
| Mean single sequence MFE | -34.70 |
| Consensus MFE | -25.23 |
| Energy contribution | -25.47 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15070971 98 - 22224390 GCUGGGAUGGGAUGCUAUGGAAGUUGGGCUCUGAGCAACUGACAUGCAAUUUAUAGCCCCGACUGGUAAUUGACAGUCUG-CUUCCUC----------CCUGCUUCCUU ((.((((.((((.((..........(((((.((((((.......)))...))).))))).(((((........))))).)-)))))))----------)).))...... ( -32.70) >DroSec_CAF1 32044 97 - 1 GUUGGGAUGGGAUGCUAUGGAAGUUGGGCUCUGAGCAACUGACAUGCAAUUUAUAGCCCCGACUGGUAAUUGACAGUCGG-CC-CCUC----------CCUGCUUCCUU ((.((((.(((.(((.(((..(((((.........)))))..))))))..........(((((((........)))))))-))-).))----------)).))...... ( -36.60) >DroSim_CAF1 18359 97 - 1 GCUGGGAUGGGAUGCUAUGGAAGUUGGGCUCUGAGCAACUGACAUGCAAUUUAUAGCCCCGACUGGUAAUUGACAGUCGG-CC-CCUC----------CCCGCUUCCUU ((.((((.(((.(((.(((..(((((.........)))))..))))))..........(((((((........)))))))-))-).))----------)).))...... ( -38.20) >DroYak_CAF1 25451 102 - 1 GCUUG-----GAUGCCAUGGAAGUUGGGCUCUGAGCAACUGACAUGCAAUUUAUAGCCCCGACUGGUAAUUGACAGUCG-UCC-CUUUCCCGCGCACUCCUGCUUCCUU ((..(-----(((((...(((((..(((((.((((((.......)))...))).)))))((((((........))))))-...-.)))))...))).))).))...... ( -31.30) >consensus GCUGGGAUGGGAUGCUAUGGAAGUUGGGCUCUGAGCAACUGACAUGCAAUUUAUAGCCCCGACUGGUAAUUGACAGUCGG_CC_CCUC__________CCUGCUUCCUU ..................((((((.(((((.((((((.......)))...))).)))))((((((........))))))......................)))))).. (-25.23 = -25.47 + 0.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:15 2006