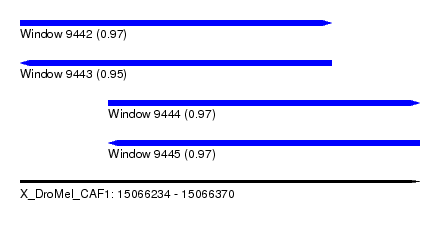
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,066,234 – 15,066,370 |
| Length | 136 |
| Max. P | 0.969128 |

| Location | 15,066,234 – 15,066,340 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.06 |
| Mean single sequence MFE | -21.57 |
| Consensus MFE | -16.23 |
| Energy contribution | -15.90 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15066234 106 + 22224390 -----CACUCGCGGUGUAAGCACACCAUCGCACCA---------UUCGCCCCAACCCACCCUUUGGCUCAGUCAUGCUCUGCGAAUUUUUCGCACAUUCCUACCCCUUUCUUUUCCAUCU -----...(((((((((....)))))...(((..(---------((.(..((((........))))..))))..)))...)))).................................... ( -16.90) >DroEre_CAF1 18364 111 + 1 ACUCGCACUCGCGGGGUAAGAACCACCUCGCACCAC--------CUCGCACC-ACCCACUUUGCAGCCCAGUCAUGCUCUGCGAAUUUUUCGCACAGUUCUACCCCUUCCUUUUCCAUGU ....((....))((((((.((((......((.....--------...))...-......(((((((..((....))..)))))))...........)))))))))).............. ( -25.70) >DroYak_CAF1 20794 114 + 1 -----CACUCGCGGGGUAAGCACCACCUCGCACCACUUGCGCCACUUGCACC-ACCCACUUUUCAGCCCAGUCAUGCUCUGCGAAUUUUUCGCACAUUUCUACCCCUUUCUUUUUCAUAU -----.......((((((.(((......((((.....)))).....)))...-...........(((........))).(((((.....)))))......)))))).............. ( -22.10) >consensus _____CACUCGCGGGGUAAGCACCACCUCGCACCAC________CUCGCACC_ACCCACUUUUCAGCCCAGUCAUGCUCUGCGAAUUUUUCGCACAUUUCUACCCCUUUCUUUUCCAUAU ..........(((((((.......))))))).................................(((........))).(((((.....))))).......................... (-16.23 = -15.90 + -0.33)


| Location | 15,066,234 – 15,066,340 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.06 |
| Mean single sequence MFE | -32.97 |
| Consensus MFE | -22.64 |
| Energy contribution | -22.87 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15066234 106 - 22224390 AGAUGGAAAAGAAAGGGGUAGGAAUGUGCGAAAAAUUCGCAGAGCAUGACUGAGCCAAAGGGUGGGUUGGGGCGAA---------UGGUGCGAUGGUGUGCUUACACCGCGAGUG----- ..............(.(((.......(((((.....)))))((((((..((.((((........)))).))(((..---------...)))......))))))..))).).....----- ( -24.80) >DroEre_CAF1 18364 111 - 1 ACAUGGAAAAGGAAGGGGUAGAACUGUGCGAAAAAUUCGCAGAGCAUGACUGGGCUGCAAAGUGGGU-GGUGCGAG--------GUGGUGCGAGGUGGUUCUUACCCCGCGAGUGCGAGU ..............((((((((((..(((((.....))))...((((.((((.((..(.......).-.)).)..)--------)).))))...)..)))).))))))((....)).... ( -37.30) >DroYak_CAF1 20794 114 - 1 AUAUGAAAAAGAAAGGGGUAGAAAUGUGCGAAAAAUUCGCAGAGCAUGACUGGGCUGAAAAGUGGGU-GGUGCAAGUGGCGCAAGUGGUGCGAGGUGGUGCUUACCCCGCGAGUG----- ..............(((((((....(..(.(....((((((..((((.(((.(.((....))).)))-.))))...(.((....)).))))))).).)..)))))))).......----- ( -36.80) >consensus ACAUGGAAAAGAAAGGGGUAGAAAUGUGCGAAAAAUUCGCAGAGCAUGACUGGGCUGAAAAGUGGGU_GGUGCGAG________GUGGUGCGAGGUGGUGCUUACCCCGCGAGUG_____ ..............(((((((...(.(((((.....))))).)((((.((...(((....)))..((....))...........)).))))..........)))))))............ (-22.64 = -22.87 + 0.22)


| Location | 15,066,264 – 15,066,370 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 87.34 |
| Mean single sequence MFE | -28.04 |
| Consensus MFE | -20.18 |
| Energy contribution | -20.34 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.967006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15066264 106 + 22224390 ----UUCGCCCCAACCCACCCUUUGGCUCAGUCAUGCUCUGCGAAUUUUUCGCACAUUCCUACCCCUUUCUUUUCCAUCUGAUAC-------CUUAGGAUGUGCUGGGGCAUGUGCA ----...((((((.........((.((..((.....))..)).))......((((((.((((......((..........))...-------..))))))))))))))))....... ( -28.10) >DroSec_CAF1 27483 105 + 1 ----UUCGCCCC-ACCCACCCUUUAGCCCAGUCAUGCUCUGCGAAUUUUUCGCACAUUCCUACCCCUUUCUUUUCCAUCUGAUAC-------CUUAGGGUGUGCUGGGGCAUGUGCA ----...(((((-(.(((((((..(((........))).(((((.....)))))...............................-------...)))))).).))))))....... ( -29.60) >DroSim_CAF1 13841 105 + 1 ----UUCGCCCC-ACCCACCCUUUAGCCCAGUCAUGCUCUGCGAAUUUUUCGCACAUUCCUACCCCUUUCUUUUCCAUCUGAUAC-------CUUAGGGUGUGCUGGGGCAUGUGCA ----...(((((-(.(((((((..(((........))).(((((.....)))))...............................-------...)))))).).))))))....... ( -29.60) >DroEre_CAF1 18400 105 + 1 ----CUCGCACC-ACCCACUUUGCAGCCCAGUCAUGCUCUGCGAAUUUUUCGCACAGUUCUACCCCUUCCUUUUCCAUGUGAUAC-------CUUAGGAUGUGCUGGGGCAUAUGCA ----...(((..-.((((.(((((((..((....))..)))))))......(((((.(((((.....(((........).))...-------..)))))))))))))).....))). ( -26.70) >DroYak_CAF1 20829 116 + 1 GCCACUUGCACC-ACCCACUUUUCAGCCCAGUCAUGCUCUGCGAAUUUUUCGCACAUUUCUACCCCUUUCUUUUUCAUAUGAUACCAUAUACCAUAGGGUGUACUGGGGCAUAUGCA ((....(((...-.....((....))((((((.(((((((((((.....)))).......................(((((....))))).....))))))))))))))))...)). ( -26.20) >consensus ____UUCGCCCC_ACCCACCCUUUAGCCCAGUCAUGCUCUGCGAAUUUUUCGCACAUUCCUACCCCUUUCUUUUCCAUCUGAUAC_______CUUAGGGUGUGCUGGGGCAUGUGCA .......(((((....((((((..(((........))).(((((.....))))).........................................))))))....)))))....... (-20.18 = -20.34 + 0.16)

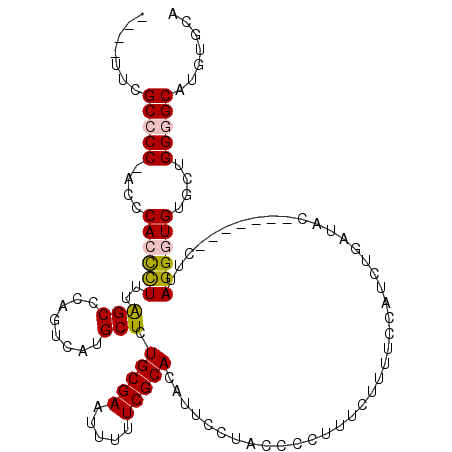
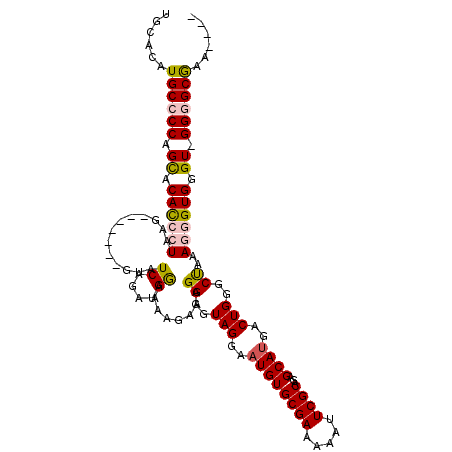
| Location | 15,066,264 – 15,066,370 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 87.34 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -25.16 |
| Energy contribution | -25.68 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15066264 106 - 22224390 UGCACAUGCCCCAGCACAUCCUAAG-------GUAUCAGAUGGAAAAGAAAGGGGUAGGAAUGUGCGAAAAAUUCGCAGAGCAUGACUGAGCCAAAGGGUGGGUUGGGGCGAA---- ......(((((((((.((((((...-------...((.....)).......((..(((..((((((((.....))))...))))..)))..))..)))))).)))))))))..---- ( -38.80) >DroSec_CAF1 27483 105 - 1 UGCACAUGCCCCAGCACACCCUAAG-------GUAUCAGAUGGAAAAGAAAGGGGUAGGAAUGUGCGAAAAAUUCGCAGAGCAUGACUGGGCUAAAGGGUGGGU-GGGGCGAA---- ......(((((((.(.((((((..(-------((.((.....))...........(((..((((((((.....))))...))))..))).)))..))))))).)-))))))..---- ( -36.90) >DroSim_CAF1 13841 105 - 1 UGCACAUGCCCCAGCACACCCUAAG-------GUAUCAGAUGGAAAAGAAAGGGGUAGGAAUGUGCGAAAAAUUCGCAGAGCAUGACUGGGCUAAAGGGUGGGU-GGGGCGAA---- ......(((((((.(.((((((..(-------((.((.....))...........(((..((((((((.....))))...))))..))).)))..))))))).)-))))))..---- ( -36.90) >DroEre_CAF1 18400 105 - 1 UGCAUAUGCCCCAGCACAUCCUAAG-------GUAUCACAUGGAAAAGGAAGGGGUAGAACUGUGCGAAAAAUUCGCAGAGCAUGACUGGGCUGCAAAGUGGGU-GGUGCGAG---- ......((((((..(...(((...(-------......)..)))....)..))))))....(.(((((.....))))).)((((.(((.(.((....))).)))-.))))...---- ( -28.70) >DroYak_CAF1 20829 116 - 1 UGCAUAUGCCCCAGUACACCCUAUGGUAUAUGGUAUCAUAUGAAAAAGAAAGGGGUAGAAAUGUGCGAAAAAUUCGCAGAGCAUGACUGGGCUGAAAAGUGGGU-GGUGCAAGUGGC .((((.((((((.....(((....)))(((((....)))))..........))))))...))))((((.....))))...((((.(((.(.((....))).)))-.))))....... ( -33.20) >consensus UGCACAUGCCCCAGCACACCCUAAG_______GUAUCAGAUGGAAAAGAAAGGGGUAGGAAUGUGCGAAAAAUUCGCAGAGCAUGACUGGGCUAAAGGGUGGGU_GGGGCGAA____ ......((((((.((.((((((.............((....))........((..(((..((((((((.....))))...))))..)))..))..)))))).)).))))))...... (-25.16 = -25.68 + 0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:09 2006