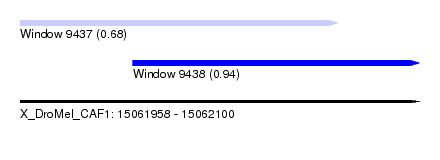
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,061,958 – 15,062,100 |
| Length | 142 |
| Max. P | 0.935130 |

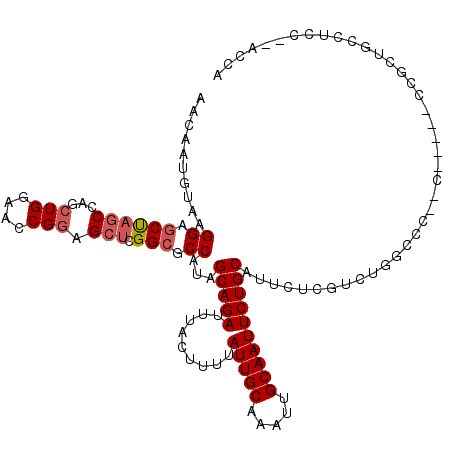
| Location | 15,061,958 – 15,062,071 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.31 |
| Mean single sequence MFE | -30.42 |
| Consensus MFE | -19.45 |
| Energy contribution | -19.98 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679155 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15061958 113 + 22224390 AACAAUGUAAGGAGCUAGCCAGCUGGAACCGGAGCUCGGCGCCAUAGCAGAUUUACUUUUAUUGCAAAUUGCAAUUCUGCAUUCUCGUCUGGCCCG----CCCCCGCUCCCUCC---CCU ..........(((((.((....))((...(((.(((.((((.....(((((.........(((((.....)))))))))).....)))).))))))----...)))))))....---... ( -32.80) >DroSec_CAF1 23227 118 + 1 AACAAUGUAAGGAGCUAGCCAGCUGGAACCGGAGCUCGGCGCCAUAGCAGAUUUUCUUUUAUUGCAAAUUGCAAUUCUGCAUUCUCGUCUGGCCCCCCAACCCCCGCUGCCACC--ACCA ..........((.(...((.(((.((....((.(((.((((.....(((((.........(((((.....)))))))))).....)))).))).))......)).)))))...)--.)). ( -28.60) >DroAna_CAF1 15897 95 + 1 AACAGUGCCAGGACCCUCCCAUCUGGAGCCG--GCUUGGCGCCACAGCAGAUUUACUUUUAUUGCAAAUUGCAAUUCUGCAUCCUCGGUUAC-----C------------------AACU (((.((((((((.((((((.....))))..)--)))))))))....(((((.........(((((.....)))))))))).......)))..-----.------------------.... ( -29.70) >DroSim_CAF1 9636 118 + 1 AACAAUGUAAGGAGCUAGCCAGCUGGAACCGGAGCUCGGCGCCAUAGCAGAUUUACUUUUAUUGCAAAUUGCAAUUCUGCAUUCUCGUCUGGCCCCCCAACCCCCGCUGCCUCC--ACCA ..........((((...((.(((.((....((.(((.((((.....(((((.........(((((.....)))))))))).....)))).))).))......)).)))))))))--.... ( -32.30) >DroEre_CAF1 14437 103 + 1 AACAAUGUAAGGAGCUAGCCAGCUGGAACCGGAGCUCGGCGCCAAAGCAGAUUUACUUUUAUUGCAAAUUGCAAUUCUGCAUUCUCGUCUG-----------------GCAAAUGCACCA ..........((.((..(((((((((..(((.....)))..)))..(((((.........(((((.....))))))))))......).)))-----------------))....)).)). ( -30.30) >DroYak_CAF1 16407 116 + 1 AACAAUGUAAGGAGCUAGCCAACUGGAACCGGAGCUCGGCGCCAUAGCAGAUUUACUUUUAUUGCAAAUUGCAAUUCUGCAUUCUCGUCUGACCCUCCA----CCGAUGCAUAUGCAACA ....(((((.((..((((....))))..))((((.((((((.....(((((.........(((((.....)))))))))).....)).))))..)))).----....)))))........ ( -28.80) >consensus AACAAUGUAAGGAGCUAGCCAGCUGGAACCGGAGCUCGGCGCCAUAGCAGAUUUACUUUUAUUGCAAAUUGCAAUUCUGCAUUCUCGUCUGGCCC__C_____CCGCUGCCUCC__ACCA ..........((.((((((...(((....))).))).))).))...(((((.........(((((.....))))))))))........................................ (-19.45 = -19.98 + 0.53)


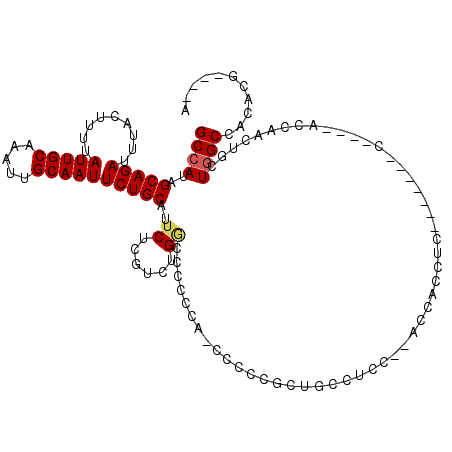
| Location | 15,061,998 – 15,062,100 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.09 |
| Mean single sequence MFE | -19.01 |
| Consensus MFE | -11.72 |
| Energy contribution | -12.00 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15061998 102 + 22224390 GCCAUAGCAGAUUUACUUUUAUUGCAAAUUGCAAUUCUGCAUUCUCGUCUGGCCCG----CCCCCGCUCCCUCC---CCUCCUC-------CGCCUACCAACUGCUGGCCACACG----A ......(((((.........(((((.....))))))))))....((((.(((((.(----(.............---.......-------............)).))))).)))----) ( -22.85) >DroSec_CAF1 23267 103 + 1 GCCAUAGCAGAUUUUCUUUUAUUGCAAAUUGCAAUUCUGCAUUCUCGUCUGGCCCCCCAACCCCCGCUGCCACC--ACCACCUC-------C----ACCAACUGCUGGCCACACG----A ......(((((.........(((((.....))))))))))....((((.(((((....................--........-------.----..........))))).)))----) ( -19.63) >DroSim_CAF1 9676 103 + 1 GCCAUAGCAGAUUUACUUUUAUUGCAAAUUGCAAUUCUGCAUUCUCGUCUGGCCCCCCAACCCCCGCUGCCUCC--ACCACCUC-------C----ACCAACUGCUGGCCACACG----A ......(((((.........(((((.....))))))))))....((((.(((((....................--........-------.----..........))))).)))----) ( -19.63) >DroEre_CAF1 14477 94 + 1 GCCAAAGCAGAUUUACUUUUAUUGCAAAUUGCAAUUCUGCAUUCUCGUCUG-----------------GCAAAUGCACCACCCCCCAACUCCC-----CCUCCGCUGGCCACAGG----A .((...(((((.........(((((.....)))))))))).........((-----------------((....((.................-----.....))..))))..))----. ( -16.95) >DroYak_CAF1 16447 102 + 1 GCCAUAGCAGAUUUACUUUUAUUGCAAAUUGCAAUUCUGCAUUCUCGUCUGACCCUCCA----CCGAUGCAUAUGCAACACCCC--------------CCCCCGCUAGCCAUACCAUAUA ((..((((............(((((.....)))))..(((((...((((.(........----).))))...))))).......--------------.....))))))........... ( -16.00) >consensus GCCAUAGCAGAUUUACUUUUAUUGCAAAUUGCAAUUCUGCAUUCUCGUCUGGCCCCCCA_CCCCCGCUGCCUCC__ACCACCUC_______C____ACCAACUGCUGGCCACACG____A ((((..(((((.........(((((.....))))))))))..((......)).....................................................))))........... (-11.72 = -12.00 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:04 2006