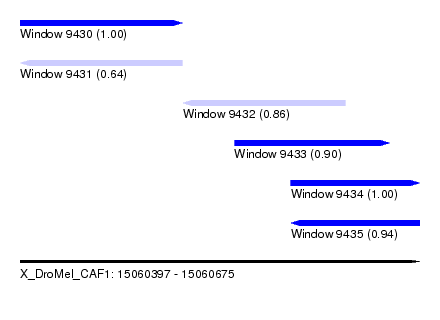
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,060,397 – 15,060,675 |
| Length | 278 |
| Max. P | 0.998051 |

| Location | 15,060,397 – 15,060,510 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.24 |
| Mean single sequence MFE | -35.36 |
| Consensus MFE | -23.98 |
| Energy contribution | -26.06 |
| Covariance contribution | 2.08 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.995954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15060397 113 + 22224390 AAAAUUUCUUUCUCUGCCCGAAUCCCGCAGAAGGGCAACCAAAUGGAAAUUGCCAGUGGUGGGGUUUUCGACGGGGGUCUUCCUUAACACAAAAAU---AA----AAAAGAGAAAGAUAU ......(((((((((.((((((((((((.....(((((((....))...)))))....)))))))).....))))((....)).............---..----...)))))))))... ( -37.00) >DroSec_CAF1 21631 119 + 1 ACAAUUUCUUUCUCUCCUCGAAUCCCGCAGAAGGGCAACUAAAUGGAAAUUGCCAGUGGUGGGGUUUUCGAGGGG-GGCUUCCUUAACACAAAAAAAAAAAAAAGAAAAGAGAAAGGUAU ......((((((((((((((((((((((.....(((((...........)))))....)))))))..))))))((-(....)))........................)))))))))... ( -37.10) >DroSim_CAF1 7498 116 + 1 ACAAUUUCUUUCUCUGCUCGAAUCCCGCAGAAGGGCAACCAAAUGGAAAUUGCCAGUGGUGGGGUUUUCGAGGGG-GGCUUCCUUAACACAAAAAA---AAAAAGAAAAGAGAAGGGUAU ......(((((((((.((((((((((((.....(((((((....))...)))))....)))))))..)))))(((-(....))))...........---.........)))))))))... ( -37.60) >DroEre_CAF1 13013 104 + 1 ACAAUUUUUUUCUCUGCUCGAAUCCCGCAGAAGGGCAACCAAAUGGAAAUUGCCAGUGGUGGGGCUU-UCGAGGGGG--UUUCUCAACACAAAA-----AGGGGGAAAA------UAC-- .....((((..((((.((((((((((((.....(((((((....))...)))))....))))))..)-)))))(((.--...))).........-----))))..))))------...-- ( -33.70) >DroYak_CAF1 14517 109 + 1 AGCAUUUUUUUCUCUGCCCGAAUCCCGCAGAAGGGCAACCGAAUGGAAAUUGCCAGUGGUGGGGUUUUUUAAGGGGG--UUCCUUGGCACAAAAAA---AAGGAGAAAA------UAUAU .......(((((((((((.(((((((((.....(((((((....))...)))))....)))))))))...((((...--..)))))))........---..))))))))------..... ( -31.40) >consensus ACAAUUUCUUUCUCUGCUCGAAUCCCGCAGAAGGGCAACCAAAUGGAAAUUGCCAGUGGUGGGGUUUUCGAAGGGGG_CUUCCUUAACACAAAAAA___AAAAAGAAAAGAGAA_GAUAU ......(((((((((....(((((((((.....(((((((....))...)))))....)))))))))...(((((.....))))).......................)))))))))... (-23.98 = -26.06 + 2.08)



| Location | 15,060,397 – 15,060,510 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.24 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -17.36 |
| Energy contribution | -17.24 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15060397 113 - 22224390 AUAUCUUUCUCUUUU----UU---AUUUUUGUGUUAAGGAAGACCCCCGUCGAAAACCCCACCACUGGCAAUUUCCAUUUGGUUGCCCUUCUGCGGGAUUCGGGCAGAGAAAGAAAUUUU ...(((((((((...----..---.(((((.(....).)))))..((((.......(((((.....(((((((.......)))))))....)).)))...)))).)))))))))...... ( -28.80) >DroSec_CAF1 21631 119 - 1 AUACCUUUCUCUUUUCUUUUUUUUUUUUUUGUGUUAAGGAAGCC-CCCCUCGAAAACCCCACCACUGGCAAUUUCCAUUUAGUUGCCCUUCUGCGGGAUUCGAGGAGAGAAAGAAAUUGU ....(((((((((((((((................)))))))..-..(((((((..(((((.....(((((((.......)))))))....)).))).)))))))))))))))....... ( -34.09) >DroSim_CAF1 7498 116 - 1 AUACCCUUCUCUUUUCUUUUU---UUUUUUGUGUUAAGGAAGCC-CCCCUCGAAAACCCCACCACUGGCAAUUUCCAUUUGGUUGCCCUUCUGCGGGAUUCGAGCAGAGAAAGAAAUUGU ................(((((---(((((((......((.....-.))((((((..(((((.....(((((((.......)))))))....)).))).)))))))))))))))))).... ( -27.00) >DroEre_CAF1 13013 104 - 1 --GUA------UUUUCCCCCU-----UUUUGUGUUGAGAAA--CCCCCUCGA-AAGCCCCACCACUGGCAAUUUCCAUUUGGUUGCCCUUCUGCGGGAUUCGAGCAGAGAAAAAAAUUGU --...------.........(-----(((((.(((....))--)...(((((-(..(((((.....(((((((.......)))))))....)).))).)))))))))))).......... ( -21.80) >DroYak_CAF1 14517 109 - 1 AUAUA------UUUUCUCCUU---UUUUUUGUGCCAAGGAA--CCCCCUUAAAAAACCCCACCACUGGCAAUUUCCAUUCGGUUGCCCUUCUGCGGGAUUCGGGCAGAGAAAAAAAUGCU .....------........((---((((((.((((((((..--...))))......(((((.....(((((((.......)))))))....)).))).....)))).))))))))..... ( -27.80) >consensus AUAUC_UUCUCUUUUCUCCUU___UUUUUUGUGUUAAGGAAG_CCCCCCUCGAAAACCCCACCACUGGCAAUUUCCAUUUGGUUGCCCUUCUGCGGGAUUCGAGCAGAGAAAGAAAUUGU .........................(((((.((((..((.......))........(((((.....(((((((.......)))))))....)).))).....)))).)))))........ (-17.36 = -17.24 + -0.12)


| Location | 15,060,510 – 15,060,623 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 92.39 |
| Mean single sequence MFE | -23.06 |
| Consensus MFE | -22.22 |
| Energy contribution | -22.26 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15060510 113 - 22224390 UGCCGCCGUUGGCUUCAUAAAAAACCAACUACAUGGAGUGCGCAUAGCAACAAA-AAACAUGCAAGCAAAUCACAGUGUACACAGCGCACUUCGCCCCCUCU----CGUUCUCAUUCU ....((.(((((.((......)).))))).....(((((((((...(((.....-.....)))..(((........))).....))))))))))).......----............ ( -24.00) >DroSec_CAF1 21750 112 - 1 CGCCGCCGUUGGCUUCAUAAAAAACCAACUACAUGGAGUGCGCAUAGCAACAAAAAAACAUGCAGGCAAAUCACAGUGUA--CAGCGCACUUCGCCCCCUCC----CGUUCUCAUUCU ....((.(((((.((......)).))))).....(((((((((...(((...........)))..(((........))).--..))))))))))).......----............ ( -24.00) >DroSim_CAF1 7614 112 - 1 CGCCGCCGUUGGCUUCAUAAAAAACCAACUACAUGGAGUGCGCAUAGCAACAAAAAAACAUGCAGGCAAAUCACAGUGUA--CAGCGCACUUCGCCCCCUCU----CGUUCUCAUUCU ....((.(((((.((......)).))))).....(((((((((...(((...........)))..(((........))).--..))))))))))).......----............ ( -24.00) >DroEre_CAF1 13117 114 - 1 CGCCGCCGUUGGCUUCAUAAAAAACCAACUACAUGGAGUGCGCAUAGCAACAA--AAACAUGCAGGCAAAUCACAGUGUA--CAGCGCCCUUCGCCCCCUCCCCCUCCUUCUCCUUCU ....((.(((((.((......)).))))).....((((.((((...(((....--.....)))..(((........))).--..)))).))))))....................... ( -19.40) >DroYak_CAF1 14626 106 - 1 CGCCGCCGUUGGCUUCAUAAAAAACCAACUACAUGGAGUGCGCAUAGCAACAA--AAACAUGCAGGCAAAUCACAGUGUA--CAGCGCACUUCACCCCC--CC------UCUCAUUCU .......(((((.((......)).)))))....((((((((((...(((....--.....)))..(((........))).--..)))))))))).....--..------......... ( -23.90) >consensus CGCCGCCGUUGGCUUCAUAAAAAACCAACUACAUGGAGUGCGCAUAGCAACAAA_AAACAUGCAGGCAAAUCACAGUGUA__CAGCGCACUUCGCCCCCUCC____CGUUCUCAUUCU .......(((((.((......)).)))))....((((((((((...(((...........)))..(((........))).....))))))))))........................ (-22.22 = -22.26 + 0.04)


| Location | 15,060,546 – 15,060,654 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 88.95 |
| Mean single sequence MFE | -31.72 |
| Consensus MFE | -25.32 |
| Energy contribution | -25.84 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.902587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15060546 108 + 22224390 CACUGUGAUUUGCUUGCAUGUUU-UUUGUUGCUAUGCGCACUCCAUGUAGUUGGUUUUUUAUGAAGCCAACGGCGGCAAGCAAAUGUAUACUACACCCCAAUUUCGGGG ...((((((((((((((......-.....(((.....))).....(((.(((((((((....))))))))).))))))))))))).)))).....((((......)))) ( -35.40) >DroSec_CAF1 21784 109 + 1 CACUGUGAUUUGCCUGCAUGUUUUUUUGUUGCUAUGCGCACUCCAUGUAGUUGGUUUUUUAUGAAGCCAACGGCGGCGAGCAAAUGUAUACUACACCCCAAUUUCGGGG .((..((.((((((.((((((.........)).))))((((.....)).(((((((((....))))))))).))))))))))...))........((((......)))) ( -31.70) >DroSim_CAF1 7648 109 + 1 CACUGUGAUUUGCCUGCAUGUUUUUUUGUUGCUAUGCGCACUCCAUGUAGUUGGUUUUUUAUGAAGCCAACGGCGGCGAGCAAAUGUAUACUACACCCCAAUUUCGGGG .((..((.((((((.((((((.........)).))))((((.....)).(((((((((....))))))))).))))))))))...))........((((......)))) ( -31.70) >DroEre_CAF1 13155 89 + 1 CACUGUGAUUUGCCUGCAUGUUU--UUGUUGCUAUGCGCACUCCAUGUAGUUGGUUUUUUAUGAAGCCAACGGCGGCGGGCAGAAGCAUAC------------------ ...(((..(((((((((......--....(((.....))).....(((.(((((((((....))))))))).)))))))))))).)))...------------------ ( -30.90) >DroYak_CAF1 14656 107 + 1 CACUGUGAUUUGCCUGCAUGUUU--UUGUUGCUAUGCGCACUCCAUGUAGUUGGUUUUUUAUGAAGCCAACGGCGGCGAGCAAAUGUAUACUACACCCCAAUCUCGCAA ..........((((((((((...--..((.((.....))))..))))))(((((((((....)))))))))))))(((((....((((...)))).......))))).. ( -28.90) >consensus CACUGUGAUUUGCCUGCAUGUUU_UUUGUUGCUAUGCGCACUCCAUGUAGUUGGUUUUUUAUGAAGCCAACGGCGGCGAGCAAAUGUAUACUACACCCCAAUUUCGGGG ...((((((((((.(((............(((.....))).....(((.(((((((((....))))))))).)))))).)))))).)))).....((((......)))) (-25.32 = -25.84 + 0.52)

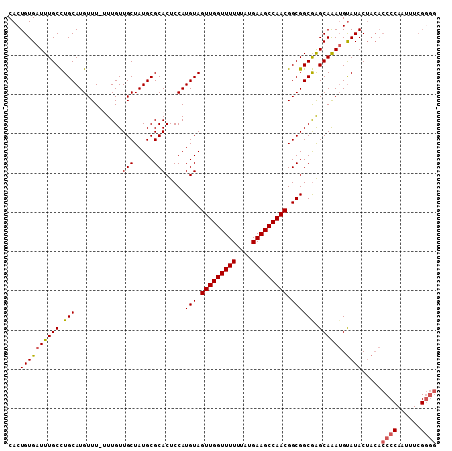
| Location | 15,060,585 – 15,060,675 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 78.41 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -23.88 |
| Energy contribution | -24.56 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.99 |
| SVM RNA-class probability | 0.998051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15060585 90 + 22224390 CUCCAUGUAGUUGGUUUUUUAUGAAGCCAACGGCGGCAAGCAAAUGUAUACUACACCCCAAUUUCGGGG----------------GAACCCGCACUU---UUCAUGGAC .((((((..(((((((((....))))))))).((((........((((...))))((((......))))----------------....))))....---..)))))). ( -32.50) >DroSec_CAF1 21824 90 + 1 CUCCAUGUAGUUGGUUUUUUAUGAAGCCAACGGCGGCGAGCAAAUGUAUACUACACCCCAAUUUCGGGG----------------GACCCCGCACUU---UUCAUGGGC .((((((..(((((((((....))))))))).((((.(..(...((((...))))((((......))))----------------)..)))))....---..)))))). ( -32.30) >DroSim_CAF1 7688 90 + 1 CUCCAUGUAGUUGGUUUUUUAUGAAGCCAACGGCGGCGAGCAAAUGUAUACUACACCCCAAUUUCGGGG----------------GACCCCGCACUU---UUCAUGGGC .((((((..(((((((((....))))))))).((((.(..(...((((...))))((((......))))----------------)..)))))....---..)))))). ( -32.30) >DroEre_CAF1 13193 74 + 1 CUCCAUGUAGUUGGUUUUUUAUGAAGCCAACGGCGGCGGGCAGAAGCAUAC---------------------------------CACCCCCACACUU--UUCCAUGGGC .((((((..(((((((((....)))))))))((.((.((((....))...)---------------------------------).)).))......--...)))))). ( -25.30) >DroYak_CAF1 14694 109 + 1 CUCCAUGUAGUUGGUUUUUUAUGAAGCCAACGGCGGCGAGCAAAUGUAUACUACACCCCAAUCUCGCAACCGCUCCCCCCACCCCACCCCCGCACUUUUUUUCAUGGGC .((((((..(((((((((....)))))))))((..(((((....((((...)))).......)))))..))...............................)))))). ( -28.10) >consensus CUCCAUGUAGUUGGUUUUUUAUGAAGCCAACGGCGGCGAGCAAAUGUAUACUACACCCCAAUUUCGGGG________________GACCCCGCACUU___UUCAUGGGC .((((((..(((((((((....))))))))).((((...((....))........((((......))))....................)))).........)))))). (-23.88 = -24.56 + 0.68)

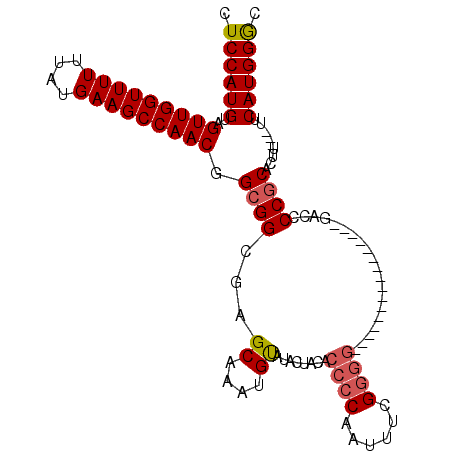
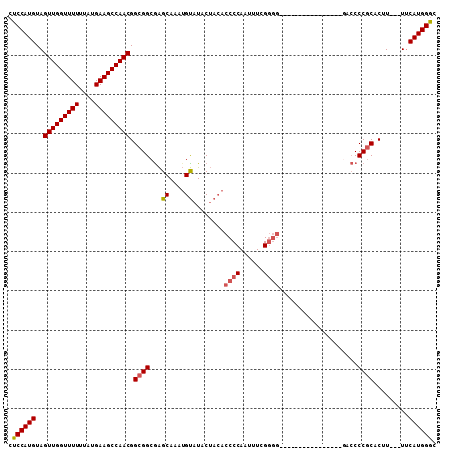
| Location | 15,060,585 – 15,060,675 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 78.41 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -21.23 |
| Energy contribution | -21.48 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15060585 90 - 22224390 GUCCAUGAA---AAGUGCGGGUUC----------------CCCCGAAAUUGGGGUGUAGUAUACAUUUGCUUGCCGCCGUUGGCUUCAUAAAAAACCAACUACAUGGAG .((((((..---..(.((((....----------------(((((....)))))...((((......))))..)))))(((((.((......)).)))))..)))))). ( -28.50) >DroSec_CAF1 21824 90 - 1 GCCCAUGAA---AAGUGCGGGGUC----------------CCCCGAAAUUGGGGUGUAGUAUACAUUUGCUCGCCGCCGUUGGCUUCAUAAAAAACCAACUACAUGGAG ..(((((..---.....((((...----------------.)))).....((((((.((((......)))))))).))(((((.((......)).)))))..))))).. ( -30.40) >DroSim_CAF1 7688 90 - 1 GCCCAUGAA---AAGUGCGGGGUC----------------CCCCGAAAUUGGGGUGUAGUAUACAUUUGCUCGCCGCCGUUGGCUUCAUAAAAAACCAACUACAUGGAG ..(((((..---.....((((...----------------.)))).....((((((.((((......)))))))).))(((((.((......)).)))))..))))).. ( -30.40) >DroEre_CAF1 13193 74 - 1 GCCCAUGGAA--AAGUGUGGGGGUG---------------------------------GUAUGCUUCUGCCCGCCGCCGUUGGCUUCAUAAAAAACCAACUACAUGGAG ..(((((...--..(.(((((((..---------------------------------(.......)..))).)))))(((((.((......)).)))))..))))).. ( -26.50) >DroYak_CAF1 14694 109 - 1 GCCCAUGAAAAAAAGUGCGGGGGUGGGGUGGGGGGAGCGGUUGCGAGAUUGGGGUGUAGUAUACAUUUGCUCGCCGCCGUUGGCUUCAUAAAAAACCAACUACAUGGAG ..(((((......(((...((..(...((((((..((((((.(((((....(((((((...))))))).))))).))))))..))))))...)..)).))).))))).. ( -36.80) >consensus GCCCAUGAA___AAGUGCGGGGUC________________CCCCGAAAUUGGGGUGUAGUAUACAUUUGCUCGCCGCCGUUGGCUUCAUAAAAAACCAACUACAUGGAG ..(((((.......(.((((.(..................(((((....)))))....(((......))).).)))))(((((.((......)).)))))..))))).. (-21.23 = -21.48 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:01 2006