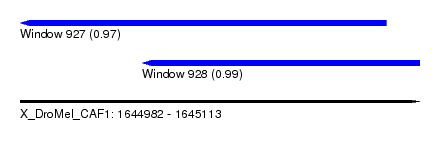
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,644,982 – 1,645,113 |
| Length | 131 |
| Max. P | 0.988943 |

| Location | 1,644,982 – 1,645,102 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.57 |
| Mean single sequence MFE | -36.98 |
| Consensus MFE | -31.80 |
| Energy contribution | -31.47 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
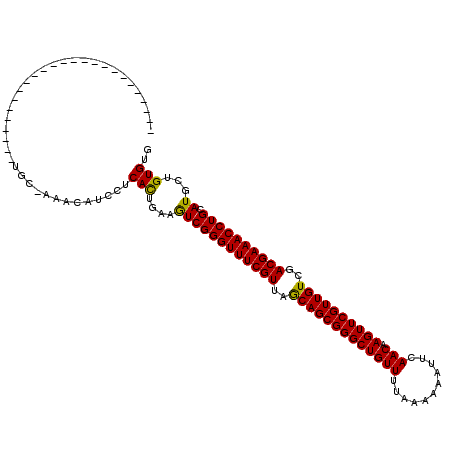
>X_DroMel_CAF1 1644982 120 - 22224390 CUGAAGUCGGGUUUCGUUAGCAGCGGGCUGUUUUAAUAAAUUCAACAAGUUCGUUGUCGACGAAACCUGCAUGCUGUGUGGAAAAUCAACGCUCAAGGCCGCCCAAGGAUUCACGUUUUU .((((..((((((((((..(((((((((((((...........))).))))))))))..)))))))))((..(((..(((.........)))....))).))....)..))))....... ( -37.40) >DroPse_CAF1 21353 91 - 1 UUGAAAUCGGGUUUCGUUAACAGCGGGCUGUUAUGAAUAAAUCAACAAGUUCGUUGUCGACGAAACCUGCAUGCUGUGUGGAGAAACAACA----------------------------- .......((((((((((..((((((((((....(((.....)))...))))))))))..))))))))))..((.(((........))).))----------------------------- ( -32.30) >DroSec_CAF1 9882 120 - 1 CUGAAGUCGGGUUUCGUUAGCAGCGGGCUGUUUUAAUAAAUUCAACAAGUUCGUUGUCGACGAAACCUGCAUGCUGUGUGGAAAAUCAACGCUCAAGGCCACUUAAGGAUCCACGUUUCU ..((((.((((((((((..(((((((((((((...........))).))))))))))..))))))))))).......(((((.......(......).((......)).)))))..))). ( -38.80) >DroEre_CAF1 11486 113 - 1 CUGAAGUCGGGUUUCGUUAGCAGCGGGCUGUUUUAAUAAAUUCAACAAGUUCGUUGUCGACGAAACCUGCAUGCUGUGUGGAAAAUCAACGCUCUAGGCCGC-CUCGGAUCUAC------ ((((.(.((((((((((..(((((((((((((...........))).))))))))))..)))))))))))..((.((.((((..........)))).)).))-.))))......------ ( -40.40) >DroYak_CAF1 10036 120 - 1 CUGAAGUCGGGUUUCGUUAGCAGCGGGCUGUUUUAAAAAAUUCAACAAGUUCGUUGUCGACGAAACCUGCAUGCUGUGUGGAAAAUCAACGCUCAAGGCCGCCCUCGGAUCUACGCUUGU .....(.((((((((((..(((((((((((((...........))).))))))))))..))))))))))).....(((((((.......(......).(((....))).))))))).... ( -40.70) >DroPer_CAF1 21944 91 - 1 UUGAAAUCGGGUUUCGUUAACAGCGGGCUGUUAUGAAUAAAUCAACAAGUUCGUUGUCGACGAAACCUGCAUGCUGUGUGGAGAAACAACA----------------------------- .......((((((((((..((((((((((....(((.....)))...))))))))))..))))))))))..((.(((........))).))----------------------------- ( -32.30) >consensus CUGAAGUCGGGUUUCGUUAGCAGCGGGCUGUUUUAAAAAAUUCAACAAGUUCGUUGUCGACGAAACCUGCAUGCUGUGUGGAAAAUCAACGCUCAAGGCCGC_C__GGAUC_AC______ .....((((((((((((..(((((((((((((...........))).))))))))))..)))))))))).))...((((.(.....).))))............................ (-31.80 = -31.47 + -0.33)

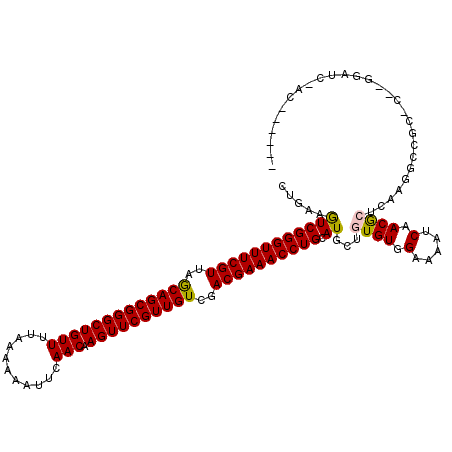
| Location | 1,645,022 – 1,645,113 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.48 |
| Mean single sequence MFE | -36.38 |
| Consensus MFE | -33.47 |
| Energy contribution | -32.80 |
| Covariance contribution | -0.67 |
| Combinations/Pair | 1.11 |
| Mean z-score | -4.04 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1645022 91 - 22224390 -----------------------------AAACAUCCUCACUGAAGUCGGGUUUCGUUAGCAGCGGGCUGUUUUAAUAAAUUCAACAAGUUCGUUGUCGACGAAACCUGCAUGCUGUGUG -----------------------------.........(((...(((((((((((((..(((((((((((((...........))).))))))))))..))))))))))...)))..))) ( -34.00) >DroPse_CAF1 21364 95 - 1 -------------------------UGCGAAAAAUCCACAUUGAAAUCGGGUUUCGUUAACAGCGGGCUGUUAUGAAUAAAUCAACAAGUUCGUUGUCGACGAAACCUGCAUGCUGUGUG -------------------------...........(((((.(....((((((((((..((((((((((....(((.....)))...))))))))))..))))))))))....).))))) ( -36.70) >DroSec_CAF1 9922 91 - 1 -----------------------------AAACAUCCUCACUGAAGUCGGGUUUCGUUAGCAGCGGGCUGUUUUAAUAAAUUCAACAAGUUCGUUGUCGACGAAACCUGCAUGCUGUGUG -----------------------------.........(((...(((((((((((((..(((((((((((((...........))).))))))))))..))))))))))...)))..))) ( -34.00) >DroEre_CAF1 11519 100 - 1 CC--------------------AACUGCAAAGCAUCCUCACUGAAGUCGGGUUUCGUUAGCAGCGGGCUGUUUUAAUAAAUUCAACAAGUUCGUUGUCGACGAAACCUGCAUGCUGUGUG ..--------------------....(((.(((((.(.....)..(.((((((((((..(((((((((((((...........))).))))))))))..)))))))))))))))).))). ( -36.50) >DroYak_CAF1 10076 100 - 1 CC--------------------AAGUGCAAAGCAUCCUCACUGAAGUCGGGUUUCGUUAGCAGCGGGCUGUUUUAAAAAAUUCAACAAGUUCGUUGUCGACGAAACCUGCAUGCUGUGUG ..--------------------....(((.(((((.(.....)..(.((((((((((..(((((((((((((...........))).))))))))))..)))))))))))))))).))). ( -36.50) >DroPer_CAF1 21955 120 - 1 CCAAAUAUUUAUUUCGAAUCUUUGAUGCGAAAAAUCCACAUUGAAAUCGGGUUUCGUUAACAGCGGGCUGUUAUGAAUAAAUCAACAAGUUCGUUGUCGACGAAACCUGCAUGCUGUGUG ...........(((((.(((...))).)))))....(((((.(....((((((((((..((((((((((....(((.....)))...))))))))))..))))))))))....).))))) ( -40.60) >consensus _________________________UGC_AAACAUCCUCACUGAAGUCGGGUUUCGUUAGCAGCGGGCUGUUUUAAAAAAUUCAACAAGUUCGUUGUCGACGAAACCUGCAUGCUGUGUG ......................................(((....((((((((((((..(((((((((((((...........))).))))))))))..)))))))))).))...))).. (-33.47 = -32.80 + -0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:29 2006