
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,047,546 – 15,047,732 |
| Length | 186 |
| Max. P | 0.937826 |

| Location | 15,047,546 – 15,047,662 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.73 |
| Mean single sequence MFE | -36.58 |
| Consensus MFE | -28.20 |
| Energy contribution | -27.92 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15047546 116 + 22224390 GGGGG-CUUUGGGC-CAAAACUUUCUAUAGCCUACAAAUUUUCUUUUGCCUCCAAAUUGGCUGAGGGAAGAAAGUUUUCCUUGCGGCUACUUUCGCCGC--UGCUCGGUCACCGAUGUCG (((((-(..(((((-..............))))).(((......)))))))))....((((((((((((((...))))))..(((((.......)))))--..))))))))......... ( -37.94) >DroSec_CAF1 3458 115 + 1 -GGGG-CUUUGGGC-CAAAACUUUCUAUAGCCUACAAAUUUUCUUUUGGCUCCAAAUUGGCUGAGGGAAGAAAGUUUUCCUUGCGGCUACUUUCGCCGC--UGCUCGGUCACCGAUGUCG -((((-(..(((((-..............)))))((((......)))))))))....((((((((((((((...))))))..(((((.......)))))--..))))))))......... ( -38.04) >DroAna_CAF1 2997 109 + 1 ---------CGGGCUGAAAACUUUCUAUAGCCUACAAAUUUUCUUUUGGCCCCAAAUUGGCUGAGGGAGGAAAGUUUUCCUUGCGGCUACUUUCGCCGCCCUGCUCGGCGACACAUCC-- ---------(((((.(((((((((((...(....)......(((((..(((.......)))..))))))))))))))))...(((((.......)))))...)))))...........-- ( -37.10) >DroEre_CAF1 3232 111 + 1 ------CUUUGGGC-CAAAACUUUCUAUAGCCUACAAAUUUUCUUUUGGCUCCAAAUUGGCUGAGGGAAGAAAGUUUUGCUUGCGGCUACUUUCGCCGC--UGCUCGGUCACCGAUGUCG ------.....(((-(((((((((((...(....)......(((((..(((.......)))..)))))))))))))))((..(((((.......)))))--.))..)))).......... ( -34.10) >DroYak_CAF1 3815 117 + 1 GGGGGCUUUUGGGC-CAAAACUUUCUAUAGCCUACAAAUUUUCUUUUGGCUCCAAAUUGGCUGAGGGAAGAAAGUUUUACUUGCGGCUACUUUCGCCGC--UGCUCGGUCACCGAUGUCG ((.((((...((((-.((((((((((...(....)......(((((..(((.......)))..)))))))))))))))....(((((.......)))))--.)))))))).))....... ( -35.70) >consensus _GGGG_CUUUGGGC_CAAAACUUUCUAUAGCCUACAAAUUUUCUUUUGGCUCCAAAUUGGCUGAGGGAAGAAAGUUUUCCUUGCGGCUACUUUCGCCGC__UGCUCGGUCACCGAUGUCG .........(((((..((((((((((...(....)......((((((((((.......))))))))))))))))))))....(((((.......)))))...)))))............. (-28.20 = -27.92 + -0.28)



| Location | 15,047,624 – 15,047,732 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 89.21 |
| Mean single sequence MFE | -25.25 |
| Consensus MFE | -20.62 |
| Energy contribution | -20.88 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.706787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15047624 108 + 22224390 UUGCGGCUACUUUCGCCGCUGCUCGGUCACCGAUGUCGCAGCACGAUGGCCUAGUCCAUCUAUAUCCACAUCUUUAUCUCCU-UUGCCAGCUCCAUUCAUCUCGACCCA ..(((((.......))))).....((((...((((..(.(((..(((((......)))))........((............-.))...))).)...))))..)))).. ( -24.42) >DroSec_CAF1 3535 108 + 1 UUGCGGCUACUUUCGCCGCUGCUCGGUCACCGAUGUCGCAGGACGAUGGCCUAGUCCAUCUAUAUCCACAUCUUUAUCUCCU-UUGCCAGCUCCAUUCAUCUCGCCCCA ..(((((.......))))).(((.(((.......(((....)))(((((......)))))......................-..)))))).................. ( -23.00) >DroEre_CAF1 3305 106 + 1 UUGCGGCUACUUUCGCCGCUGCUCGGUCACCGAUGUCGCAGCCAGAUGGCCAAGUCCAUCUAUAUCCACAUCUUUAUCUGCU-UGGCAAGCACCAUGCCUCCCCUCC-- ..(((((.......)))(((((((((...))))....))))).((((((......))))))..................)).-.((((.......))))........-- ( -26.90) >DroYak_CAF1 3894 107 + 1 UUGCGGCUACUUUCGCCGCUGCUCGGUCACCGAUGUCGCAGCAAGAUGGCCUAGUCCAUCUAUAUCCACAUCUUUAUCUCCUCUAGCCAGCUUCGUUCCCCUCUUUC-- ..(((((((........(((((((((...))))....))))).((((((......))))))......................))))).))................-- ( -26.70) >consensus UUGCGGCUACUUUCGCCGCUGCUCGGUCACCGAUGUCGCAGCAAGAUGGCCUAGUCCAUCUAUAUCCACAUCUUUAUCUCCU_UUGCCAGCUCCAUUCAUCUCGUCC__ ..(((((.......)))(((((((((...))))....)))))..(((((......))))).............................)).................. (-20.62 = -20.88 + 0.25)


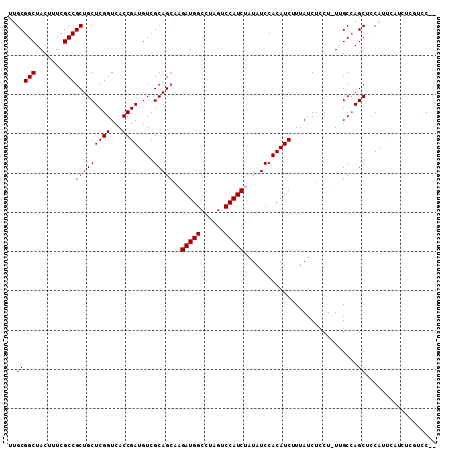
| Location | 15,047,624 – 15,047,732 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 89.21 |
| Mean single sequence MFE | -34.88 |
| Consensus MFE | -29.58 |
| Energy contribution | -29.57 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15047624 108 - 22224390 UGGGUCGAGAUGAAUGGAGCUGGCAA-AGGAGAUAAAGAUGUGGAUAUAGAUGGACUAGGCCAUCGUGCUGCGACAUCGGUGACCGAGCAGCGGCGAAAGUAGCCGCAA ..(((((...(((.((..((.((((.-.....(((....))).......(((((......))))).))))))..))))).))))).....(((((.......))))).. ( -33.60) >DroSec_CAF1 3535 108 - 1 UGGGGCGAGAUGAAUGGAGCUGGCAA-AGGAGAUAAAGAUGUGGAUAUAGAUGGACUAGGCCAUCGUCCUGCGACAUCGGUGACCGAGCAGCGGCGAAAGUAGCCGCAA ((.(((............((((.(..-.((..((...(((((.(.....(((((......)))))......).))))).))..))..))))).((....)).))).)). ( -33.40) >DroEre_CAF1 3305 106 - 1 --GGAGGGGAGGCAUGGUGCUUGCCA-AGCAGAUAAAGAUGUGGAUAUAGAUGGACUUGGCCAUCUGGCUGCGACAUCGGUGACCGAGCAGCGGCGAAAGUAGCCGCAA --....((.((((.....)))).)).-.((.......(((((.(...(((((((......)))))))....).)))))(....)...)).(((((.......))))).. ( -39.90) >DroYak_CAF1 3894 107 - 1 --GAAAGAGGGGAACGAAGCUGGCUAGAGGAGAUAAAGAUGUGGAUAUAGAUGGACUAGGCCAUCUUGCUGCGACAUCGGUGACCGAGCAGCGGCGAAAGUAGCCGCAA --....................(((...((..((...(((((.(....((((((......)))))).....).))))).))..)).))).(((((.......))))).. ( -32.60) >consensus __GGACGAGAGGAAUGGAGCUGGCAA_AGGAGAUAAAGAUGUGGAUAUAGAUGGACUAGGCCAUCGUGCUGCGACAUCGGUGACCGAGCAGCGGCGAAAGUAGCCGCAA ..................(((.(..............(((((.(.....(((((......)))))......).)))))(....)).))).(((((.......))))).. (-29.58 = -29.57 + -0.00)


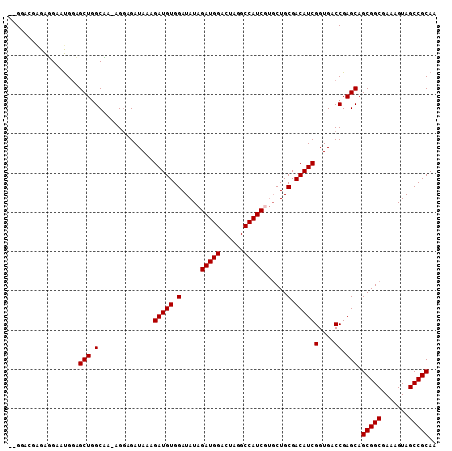
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:44 2006