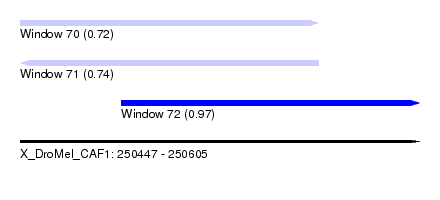
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 250,447 – 250,605 |
| Length | 158 |
| Max. P | 0.965304 |

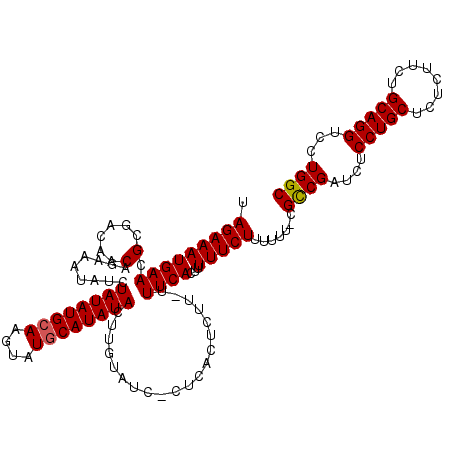
| Location | 250,447 – 250,565 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.78 |
| Mean single sequence MFE | -23.13 |
| Consensus MFE | -17.95 |
| Energy contribution | -18.07 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 250447 118 + 22224390 UAGAAAUGAACGCGACAACAGAAAUAUCUAUAUGCAAGUAUACAUAUACUUGUAUCCCUCACUCUU-UUUCACUUUUUCUUUUUU-CGCCGAUCUCCUGCUCUCUUCUGCAGGUCCUGGC ..((((.(((.........(((....)))..((((((((((....))))))))))...........-.........)))...)))-)((((....(((((........)))))...)))) ( -24.70) >DroSim_CAF1 19195 118 + 1 UAGAAAUGAACGCGACAACAGAAAUAUCUAUAUGCAAGUAUGCAUAUACUUGUAUCUCUCACUCUU-UUUCACUUUUUCUUUUUU-CGCCGAUCUCCUGCUCUCUUCUGCAGGUCCUGGC .(((((((((...((....(((.(((..((((((((....))))))))..))).))).....))..-.))))...))))).....-.((((....(((((........)))))...)))) ( -26.60) >DroYak_CAF1 19996 112 + 1 UAGAAAUGAACGCAACAACAAAAAUAUCUAUAUGCAAGUAUGCAUAUAC--------CUUACUCUUUUUUCACCUUUUCUUUUUUCCGUCGAUCUCCUGCUCUCUUCUGCAGGUCCUGGC .(((((((((.(......).........((((((((....)))))))).--------...........))))...))))).......((((....(((((........)))))...)))) ( -18.10) >consensus UAGAAAUGAACGCGACAACAGAAAUAUCUAUAUGCAAGUAUGCAUAUACUUGUAUC_CUCACUCUU_UUUCACUUUUUCUUUUUU_CGCCGAUCUCCUGCUCUCUUCUGCAGGUCCUGGC .(((((((((.(......).........((((((((....))))))))....................))))...))))).......((((....(((((........)))))...)))) (-17.95 = -18.07 + 0.11)

| Location | 250,447 – 250,565 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.78 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -24.16 |
| Energy contribution | -23.83 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 250447 118 - 22224390 GCCAGGACCUGCAGAAGAGAGCAGGAGAUCGGCG-AAAAAAGAAAAAGUGAAA-AAGAGUGAGGGAUACAAGUAUAUGUAUACUUGCAUAUAGAUAUUUCUGUUGUCGCGUUCAUUUCUA (((....(((((........))))).....)))(-(((...(((...((((..-....(..((..(((((......))))).))..)..(((((....)))))..)))).))).)))).. ( -29.40) >DroSim_CAF1 19195 118 - 1 GCCAGGACCUGCAGAAGAGAGCAGGAGAUCGGCG-AAAAAAGAAAAAGUGAAA-AAGAGUGAGAGAUACAAGUAUAUGCAUACUUGCAUAUAGAUAUUUCUGUUGUCGCGUUCAUUUCUA (((....(((((........))))).....))).-.....((((...(((((.-..((...(((((((....((((((((....))))))))..)))))))....))...))))))))). ( -30.90) >DroYak_CAF1 19996 112 - 1 GCCAGGACCUGCAGAAGAGAGCAGGAGAUCGACGGAAAAAAGAAAAGGUGAAAAAAGAGUAAG--------GUAUAUGCAUACUUGCAUAUAGAUAUUUUUGUUGUUGCGUUCAUUUCUA .......(((((........)))))........(((((...(((...(..(...(((((((..--------.((((((((....))))))))..)))))))....)..).))).))))). ( -25.10) >consensus GCCAGGACCUGCAGAAGAGAGCAGGAGAUCGGCG_AAAAAAGAAAAAGUGAAA_AAGAGUGAG_GAUACAAGUAUAUGCAUACUUGCAUAUAGAUAUUUCUGUUGUCGCGUUCAUUUCUA (((....(((((........))))).....))).......((((...(((((......((((..........((((((((....))))))))((((....)))).)))).))))))))). (-24.16 = -23.83 + -0.33)

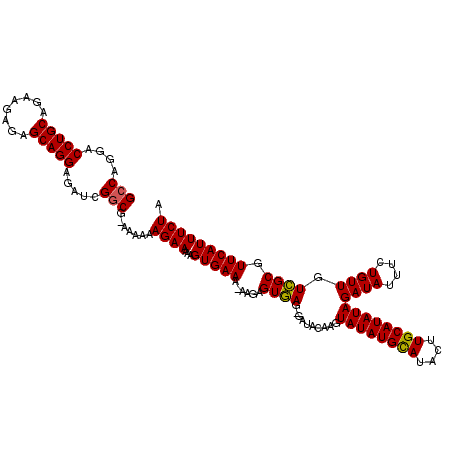
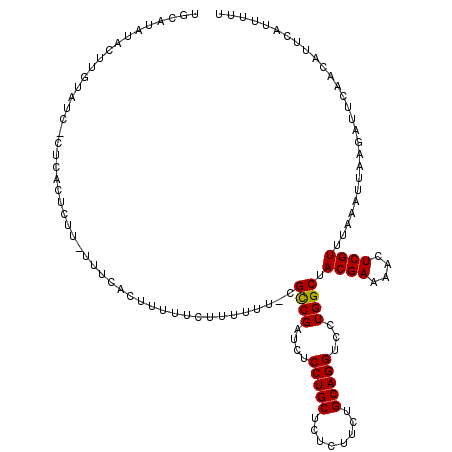
| Location | 250,487 – 250,605 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.34 |
| Mean single sequence MFE | -16.80 |
| Consensus MFE | -15.22 |
| Energy contribution | -15.00 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 250487 118 + 22224390 UACAUAUACUUGUAUCCCUCACUCUU-UUUCACUUUUUCUUUUUU-CGCCGAUCUCCUGCUCUCUUCUGCAGGUCCUGGCUACGAAAACUCGUUUAAAUUAAGAUUCAACAUUCAUUUUU ((((......))))............-..........((((..((-.((((....(((((........)))))...)))).((((....))))...))..))))................ ( -18.40) >DroSim_CAF1 19235 118 + 1 UGCAUAUACUUGUAUCUCUCACUCUU-UUUCACUUUUUCUUUUUU-CGCCGAUCUCCUGCUCUCUUCUGCAGGUCCUGGCUACGAAAACUCGUUUAAAUUAAGAUUCAACAUUCAUUUUU .........(((.((((.........-..................-.((((....(((((........)))))...)))).((((....))))........)))).)))........... ( -18.80) >DroYak_CAF1 20036 112 + 1 UGCAUAUAC--------CUUACUCUUUUUUCACCUUUUCUUUUUUCCGUCGAUCUCCUGCUCUCUUCUGCAGGUCCUGGCUACGAAAACUCGUUUAAAUUAACAUUCAACAUUCAUUUUU .........--------..............................((((....(((((........)))))...)))).((((....))))........................... ( -13.20) >consensus UGCAUAUACUUGUAUC_CUCACUCUU_UUUCACUUUUUCUUUUUU_CGCCGAUCUCCUGCUCUCUUCUGCAGGUCCUGGCUACGAAAACUCGUUUAAAUUAAGAUUCAACAUUCAUUUUU ...............................................((((....(((((........)))))...)))).((((....))))........................... (-15.22 = -15.00 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:36 2006