
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,026,950 – 15,027,081 |
| Length | 131 |
| Max. P | 0.969416 |

| Location | 15,026,950 – 15,027,057 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.97 |
| Mean single sequence MFE | -36.38 |
| Consensus MFE | -23.08 |
| Energy contribution | -24.45 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.731049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15026950 107 + 22224390 GUGGCGGGGCGGGGCGGUAUGGUGAGGUGCGGUGCUAAUUUCAUCGCCGGACACGUUUCCCCCCGGCCAAAAUGGCCAAAAAUCAAAUUGAGGA-------GAACCGGAAACGG .....((((.(..(((((.((((((..((..((....))..)))))))).)).)))..).))))((((.....)))).................-------...(((....))) ( -40.40) >DroSec_CAF1 44466 87 + 1 GGGGCGGGGC--------------------GGUGCUAAUUUCAUCGCCGGCCACGUUUCCCCCCGGCCAAAAUGGCCAAAAAUCAAAUUGAGGA-------GAACCGGAAACGG ((((.(((((--------------------((((.......)))))))(....)....))))))((((.....)))).................-------...(((....))) ( -34.60) >DroSim_CAF1 47956 87 + 1 GGGGCGGGGC--------------------GGUGCUAAUUUCAUCGCCGGCCACGUUUCCCCCCGGCCAAAAUGGCCAAAAAUCAAAUUGAGGA-------GAACCGGAAACGG ((((.(((((--------------------((((.......)))))))(....)....))))))((((.....)))).................-------...(((....))) ( -34.60) >DroEre_CAF1 45368 99 + 1 GGGGCGG----------UUUGG-----AGCGGAGCUAAUUUCAUCGCCGGCCACGUUUUCCUCCGGCCAAAAUGGCCAAAAAUCAAAUUGGUGGGGGGUGGGAACCGGAAACGG ..(((((----------(....-----(((...)))......))))))..((((.....(((((((((.....))))....(((.....))))))))))))...(((....))) ( -35.90) >consensus GGGGCGGGGC____________________GGUGCUAAUUUCAUCGCCGGCCACGUUUCCCCCCGGCCAAAAUGGCCAAAAAUCAAAUUGAGGA_______GAACCGGAAACGG ((((.(((....................((((((.......))))))((....))..)))))))((((.....))))...........................(((....))) (-23.08 = -24.45 + 1.37)



| Location | 15,026,990 – 15,027,081 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.29 |
| Mean single sequence MFE | -33.78 |
| Consensus MFE | -21.46 |
| Energy contribution | -21.78 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15026990 91 + 22224390 UCAUCGCCGGACACGUUUCCCCCCGGCCAAAAUGGCCAAAAAUCAAAUUGAGGA------------GAACCGGAAACGGAAGCAUUGAAGAAGUGAGGG-------CAG---------G .....(((...(((.(((((....((((.....)))).....((.....)))))------------)).(((....))).............)))..))-------)..---------. ( -27.00) >DroSec_CAF1 44486 91 + 1 UCAUCGCCGGCCACGUUUCCCCCCGGCCAAAAUGGCCAAAAAUCAAAUUGAGGA------------GAACCGGAAACGGAAGCAUUGAAGAGGUGGGGG-------CAG---------G .....(((..((((.(((((....((((.....)))).....((.....)))))------------)).(((....))).............)))).))-------)..---------. ( -30.20) >DroSim_CAF1 47976 91 + 1 UCAUCGCCGGCCACGUUUCCCCCCGGCCAAAAUGGCCAAAAAUCAAAUUGAGGA------------GAACCGGAAACGGAAGCAUUGAAGAGGUGGGGG-------CAG---------G .....(((..((((.(((((....((((.....)))).....((.....)))))------------)).(((....))).............)))).))-------)..---------. ( -30.20) >DroEre_CAF1 45393 105 + 1 UCAUCGCCGGCCACGUUUUCCUCCGGCCAAAAUGGCCAAAAAUCAAAUUGGUGGGGG-----GUGGGAACCGGAAACGGAAGCCGUAAAGGAGUGGUGGAGUGGUGGAG---------G ((((((((.(((((....((((((((((.....))))....(((.....))))))))-----)..((..(((....)))...))........))))).).)))))))..---------. ( -44.80) >DroYak_CAF1 45783 119 + 1 UCAUCGCCGGCCACGUUUCCCUCUGGCCAAAAUGGCCAAAAAUCAAAUUAAGGGGAACCGGAACCGGAACCGGAAACGGAAGCAGUGAAGGAGUGUGGGUGUGAGGGAAGAAGAACAAG (((.((((.((.((.(((((((.(((((.....)))))............)))))))(((....)))..(((....))).............)))).)))))))............... ( -36.72) >consensus UCAUCGCCGGCCACGUUUCCCCCCGGCCAAAAUGGCCAAAAAUCAAAUUGAGGA____________GAACCGGAAACGGAAGCAUUGAAGAAGUGGGGG_______CAG_________G .......((....))....(((((((((.....))))................................(((....)))...............))))).................... (-21.46 = -21.78 + 0.32)

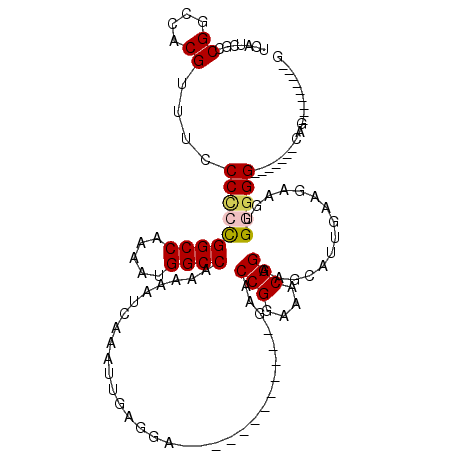

| Location | 15,026,990 – 15,027,081 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.29 |
| Mean single sequence MFE | -29.06 |
| Consensus MFE | -21.96 |
| Energy contribution | -21.76 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15026990 91 - 22224390 C---------CUG-------CCCUCACUUCUUCAAUGCUUCCGUUUCCGGUUC------------UCCUCAAUUUGAUUUUUGGCCAUUUUGGCCGGGGGGAAACGUGUCCGGCGAUGA .---------.((-------((..(((.............(((....)))(((------------(((((((........))((((.....))))))))))))..)))...)))).... ( -26.40) >DroSec_CAF1 44486 91 - 1 C---------CUG-------CCCCCACCUCUUCAAUGCUUCCGUUUCCGGUUC------------UCCUCAAUUUGAUUUUUGGCCAUUUUGGCCGGGGGGAAACGUGGCCGGCGAUGA .---------.((-------((.((((.............(((....)))(((------------(((((((........))((((.....))))))))))))..))))..)))).... ( -28.40) >DroSim_CAF1 47976 91 - 1 C---------CUG-------CCCCCACCUCUUCAAUGCUUCCGUUUCCGGUUC------------UCCUCAAUUUGAUUUUUGGCCAUUUUGGCCGGGGGGAAACGUGGCCGGCGAUGA .---------.((-------((.((((.............(((....)))(((------------(((((((........))((((.....))))))))))))..))))..)))).... ( -28.40) >DroEre_CAF1 45393 105 - 1 C---------CUCCACCACUCCACCACUCCUUUACGGCUUCCGUUUCCGGUUCCCAC-----CCCCCACCAAUUUGAUUUUUGGCCAUUUUGGCCGGAGGAAAACGUGGCCGGCGAUGA .---------........................(((((..((((((((((....))-----)................(((((((.....))))))))).))))).)))))....... ( -27.10) >DroYak_CAF1 45783 119 - 1 CUUGUUCUUCUUCCCUCACACCCACACUCCUUCACUGCUUCCGUUUCCGGUUCCGGUUCCGGUUCCCCUUAAUUUGAUUUUUGGCCAUUUUGGCCAGAGGGAAACGUGGCCGGCGAUGA ...............................(((.(((((((((((((((..(((....)))..))............((((((((.....))))))))))))))).))..)))).))) ( -35.00) >consensus C_________CUG_______CCCCCACUUCUUCAAUGCUUCCGUUUCCGGUUC____________UCCUCAAUUUGAUUUUUGGCCAUUUUGGCCGGGGGGAAACGUGGCCGGCGAUGA .........................................((((.((((((..........................((((((((.....))))))))(....)..)))))).)))). (-21.96 = -21.76 + -0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:31 2006