| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,026,065 – 15,026,180 |
| Length | 115 |
| Max. P | 0.963194 |

| Location | 15,026,065 – 15,026,180 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 84.89 |
| Mean single sequence MFE | -25.68 |
| Consensus MFE | -18.40 |
| Energy contribution | -18.52 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15026065 115 + 22224390 AUUUGCGUGCAAAAACUAGAACAACAUUUUCACAAUGGUUUUAUCCCUAU---GUAUGCCAUUUUUGCUUUAUCAGUAUAAAGCAAUUGACUAAUUAAGCAGAUUGCAUGCAGGCCCC .(((((((((((.....................((((((..(((......---))).))))))(((((((.((.(((...((....)).))).)).)))))))))))))))))).... ( -27.20) >DroSec_CAF1 43588 113 + 1 AUUUGCAUGCAAAAGCUAGGACAACAUU-CCACAAUGGUUUUAUCCCUAU---GCAUGCUAUUUUUGCUUUUCCAUAAUAAAGCAAUUGACUAAUUAAGCAGAUUGCAUGCAG-CCUC ..((((((((((..((((((........-((.....)).......)))).---)).(((((((.(((((((........)))))))......)))..))))..))))))))))-.... ( -28.26) >DroSim_CAF1 47086 113 + 1 AUUUGCAUGCAAAAGCUAGGACAACAUU-CCACAAUGGUUUUAUCCCUAU---GCAUGCUAUUUUUGCUUUCCCAGAAUAAAGCAAUUGACUAAUUAAGCAGAUUGCAUGCAG-CCCC ..((((((((((..((((((........-((.....)).......)))).---)).(((((((.(((((((........)))))))......)))..))))..))))))))))-.... ( -27.46) >DroEre_CAF1 44535 116 + 1 AGUUGCUUGCAAAUACUAAA-CAACAUU-CCACAAAGAUUUUGCCCAUAUAGUGUACAUCCUUUUUUCUUUUCCCGAAUAAAGCAAUUGACUAAUUAAGCAGAUUGCAUGCAAGCCCC ....(((((((.((((((..-...((.(-(......))...))......))))))...........................((((((..((.....))..)))))).)))))))... ( -19.80) >consensus AUUUGCAUGCAAAAACUAGAACAACAUU_CCACAAUGGUUUUAUCCCUAU___GCAUGCCAUUUUUGCUUUUCCAGAAUAAAGCAAUUGACUAAUUAAGCAGAUUGCAUGCAG_CCCC ..((((((((((..(((................((((((..................)))))).(((((((........)))))))...........)))...))))))))))..... (-18.40 = -18.52 + 0.12)



| Location | 15,026,065 – 15,026,180 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 84.89 |
| Mean single sequence MFE | -29.23 |
| Consensus MFE | -22.27 |
| Energy contribution | -22.90 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.839483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15026065 115 - 22224390 GGGGCCUGCAUGCAAUCUGCUUAAUUAGUCAAUUGCUUUAUACUGAUAAAGCAAAAAUGGCAUAC---AUAGGGAUAAAACCAUUGUGAAAAUGUUGUUCUAGUUUUUGCACGCAAAU ......(((.(((((...(((......((((.(((((((((....)))))))))...))))....---...(((((((...((((.....))))))))))))))..))))).)))... ( -27.30) >DroSec_CAF1 43588 113 - 1 GAGG-CUGCAUGCAAUCUGCUUAAUUAGUCAAUUGCUUUAUUAUGGAAAAGCAAAAAUAGCAUGC---AUAGGGAUAAAACCAUUGUGG-AAUGUUGUCCUAGCUUUUGCAUGCAAAU ....-.(((((((((...(((...........(((((((........)))))))..(((((((.(---((((((......)).))))).-.)))))))...)))..)))))))))... ( -31.90) >DroSim_CAF1 47086 113 - 1 GGGG-CUGCAUGCAAUCUGCUUAAUUAGUCAAUUGCUUUAUUCUGGGAAAGCAAAAAUAGCAUGC---AUAGGGAUAAAACCAUUGUGG-AAUGUUGUCCUAGCUUUUGCAUGCAAAU ....-.(((((((((...(((...........(((((((........)))))))..(((((((.(---((((((......)).))))).-.)))))))...)))..)))))))))... ( -32.00) >DroEre_CAF1 44535 116 - 1 GGGGCUUGCAUGCAAUCUGCUUAAUUAGUCAAUUGCUUUAUUCGGGAAAAGAAAAAAGGAUGUACACUAUAUGGGCAAAAUCUUUGUGG-AAUGUUG-UUUAGUAUUUGCAAGCAACU (..((((((((((....((((((..((((....(.((((.(((.......))).)))).).....))))..))))))...(((....))-)......-....)))..)))))))..). ( -25.70) >consensus GGGG_CUGCAUGCAAUCUGCUUAAUUAGUCAAUUGCUUUAUUCUGGAAAAGCAAAAAUAGCAUAC___AUAGGGAUAAAACCAUUGUGG_AAUGUUGUCCUAGCUUUUGCAUGCAAAU ......(((((((((...(((......(((..(((((((........)))))))....)))..........(((((((..((.....)).....))))))))))..)))))))))... (-22.27 = -22.90 + 0.63)

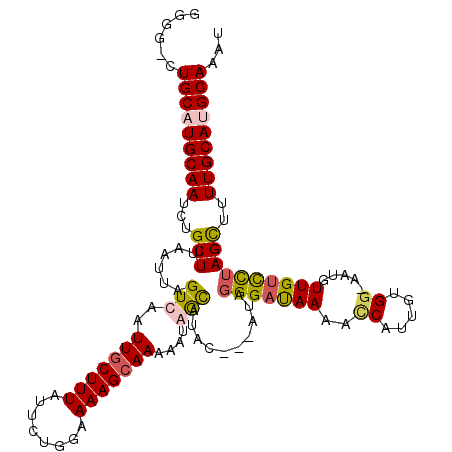

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:27 2006