
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,012,490 – 15,012,676 |
| Length | 186 |
| Max. P | 0.939480 |

| Location | 15,012,490 – 15,012,605 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 84.06 |
| Mean single sequence MFE | -50.88 |
| Consensus MFE | -40.09 |
| Energy contribution | -43.53 |
| Covariance contribution | 3.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15012490 115 + 22224390 UGCUGCCUGGGCGGCGCCCACAGUGGGUGGUGCUGCGUCAGCGGUGGUGGCGGGGCAAAAGCAGUCGCAUCACCAAUACCACCACUUGGAAAUCGGAGUGCAGGGCAAUACGCCA .(((((....)))))((((......((((((((((...))))(((((((.(((.((....))..))))))))))...))))))((((.(....).))))...))))......... ( -52.20) >DroSec_CAF1 30079 115 + 1 UGCUGCCUGGGCGGCGCCCACAGUGGGUGGUGCUGCGCCAGCGCCGGUGGUGGGGCAGAAACAGUCGCACCACCAUUACCACCACUUGGAAAUCGGAGUGCAGGGCAAUACGCCA .(((((....)))))((((...((((..(((((((...)))))))(((((((.(((.......))).)))))))....))))(((((.(....).)))))..))))......... ( -59.60) >DroEre_CAF1 31123 115 + 1 UGCUGCCUGGGGGGCGCCUACACUGGGUGGUGCAGCGGAAGCGGCGGUGGUGGGGCAGAAGCAGUCGCACCACCAUUACCACCAUUUGGAAAUCGGAGUGCUGGGCAAUACGCCU .((.(((.....)))(((((((((.((((((((.((....)).))(((((((.(((.......))).)))))))...)))))).....(....)..)))).))))).....)).. ( -53.30) >DroYak_CAF1 30995 103 + 1 UGCCGCUUGGGCGGCGCCAACAGUGGGUGGUGCAGCGGCAGC------------ACAGAAGCAGCCACACCACAAUUACCACCAUUUGGAAUUCGGAGUGCAGGGCAAUACGCCC .(((((....))))).((((..((((((((((....(((.((------------......)).))).)))))).....))))...)))).............((((.....)))) ( -38.40) >consensus UGCUGCCUGGGCGGCGCCCACAGUGGGUGGUGCAGCGGCAGCGGCGGUGGUGGGGCAGAAGCAGUCGCACCACCAUUACCACCACUUGGAAAUCGGAGUGCAGGGCAAUACGCCA .(((((....)))))((((......(((((((((((....)))))(((((((.(((.......))).)))))))...))))))((((.(....).))))...))))......... (-40.09 = -43.53 + 3.44)


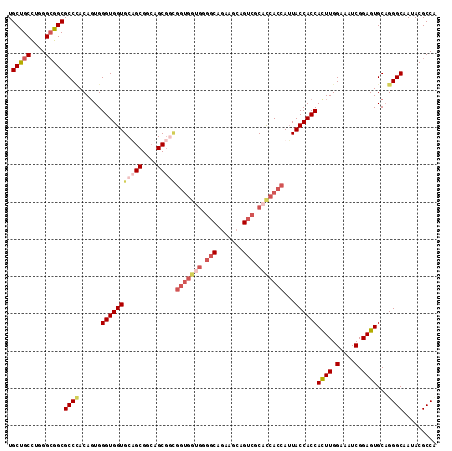
| Location | 15,012,527 – 15,012,642 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 84.49 |
| Mean single sequence MFE | -41.50 |
| Consensus MFE | -30.15 |
| Energy contribution | -33.15 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.803854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15012527 115 + 22224390 UCAGCGGUGGUGGCGGGGCAAAAGCAGUCGCAUCACCAAUACCACCACUUGGAAAUCGGAGUGCAGGGCAAUACGCCAACGUUGAUGGAGGCGAGCACCACUAGAGGUGGGAAAC .....(((((((.(((.((....))..))))))))))....(((((((((.(....).))))...((((....((((..((....))..)))).)).))......)))))..... ( -39.70) >DroSec_CAF1 30116 115 + 1 CCAGCGCCGGUGGUGGGGCAGAAACAGUCGCACCACCAUUACCACCACUUGGAAAUCGGAGUGCAGGGCAAUACGCCAACGCUGAUGGAGGCAAGCACCACCAGAGAUGGGAAAC ...((((((((((((.(((.......))).)))))))....((.((....))..(((((.((....(((.....))).)).))))))).)))..))....(((....)))..... ( -41.40) >DroEre_CAF1 31160 115 + 1 GAAGCGGCGGUGGUGGGGCAGAAGCAGUCGCACCACCAUUACCACCAUUUGGAAAUCGGAGUGCUGGGCAAUACGCCUACGCUGAUGGAGGCGAGCACCGCCAGAGGUGGGAAGC ...((..((((((((.(((.......))).)))))))....(((((.(((((....(((..((((((((.....)))).((((......))))))))))))))))))))))..)) ( -50.40) >DroYak_CAF1 31032 103 + 1 GCAGC------------ACAGAAGCAGCCACACCACAAUUACCACCAUUUGGAAUUCGGAGUGCAGGGCAAUACGCCCACGCUGAUGGAGGCGAGCACCACCAGAGGUGGGAAAC ((.((------------......)).)).............(((((.(((((......(.((((.((((.....)))).((((......)))).))))).))))))))))..... ( -34.50) >consensus GCAGCGGCGGUGGUGGGGCAGAAGCAGUCGCACCACCAUUACCACCACUUGGAAAUCGGAGUGCAGGGCAAUACGCCAACGCUGAUGGAGGCGAGCACCACCAGAGGUGGGAAAC ........(((((((.(((.......))).)))))))....(((((.(((((......(.((((..(((.....)))..((((......)))).))))).))))))))))..... (-30.15 = -33.15 + 3.00)

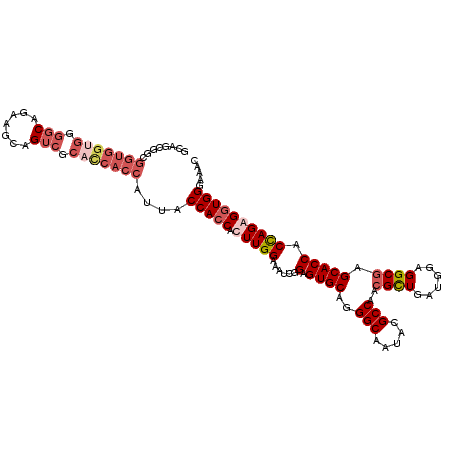
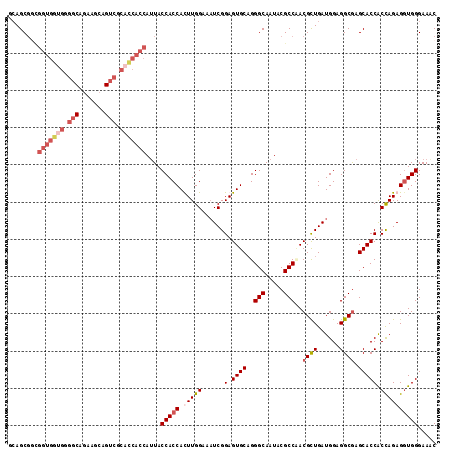
| Location | 15,012,565 – 15,012,676 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 86.26 |
| Mean single sequence MFE | -35.35 |
| Consensus MFE | -24.24 |
| Energy contribution | -23.18 |
| Covariance contribution | -1.06 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15012565 111 + 22224390 AUACCACCACUUGGAAAUCGGAGUGCAGGGCAAUACGCCAACGUUGAUGGAGGCGAGCACCACUAGAGGUGGGAAACAUGCCACAGUGCCCACCA------CUCAGGCACCACCAAA ..........((((......(((((..(((((.....(((.......))).((((..((((......))))(....).))))....)))))..))------))).(....).)))). ( -33.50) >DroSec_CAF1 30154 111 + 1 UUACCACCACUUGGAAAUCGGAGUGCAGGGCAAUACGCCAACGCUGAUGGAGGCAAGCACCACCAGAGAUGGGAAACAUGCCACAGUGGCCAUCA------CUCAGGCACCACCAAA ...(((.....))).....((.((((..(((.....)))....(((((((.((......)).)))..(((((....)..(((.....))))))).------.))))))))..))... ( -31.90) >DroEre_CAF1 31198 111 + 1 UUACCACCAUUUGGAAAUCGGAGUGCUGGGCAAUACGCCUACGCUGAUGGAGGCGAGCACCGCCAGAGGUGGGAAGCAUGGAACAGUGGCCACCA------CUCAGGCACCACCAAA ...(((((.(((((....(((..((((((((.....)))).((((......)))))))))))))))))))))...((.((....(((((...)))------)))).))......... ( -36.90) >DroYak_CAF1 31058 117 + 1 UUACCACCAUUUGGAAUUCGGAGUGCAGGGCAAUACGCCCACGCUGAUGGAGGCGAGCACCACCAGAGGUGGGAAACAUGCAAAAGUGGAUACAGUGGUGCUACAGGCAACAUCAAA ...(((((.(((((......(.((((.((((.....)))).((((......)))).))))).))))))))))......(((....((((.(((....)))))))..)))........ ( -39.10) >consensus UUACCACCACUUGGAAAUCGGAGUGCAGGGCAAUACGCCAACGCUGAUGGAGGCGAGCACCACCAGAGGUGGGAAACAUGCAACAGUGGCCACCA______CUCAGGCACCACCAAA ...((..(((((.(....).)))))..)).......(((....(((.(((..(....).))).))).(((((...((........))..)))))...........)))......... (-24.24 = -23.18 + -1.06)


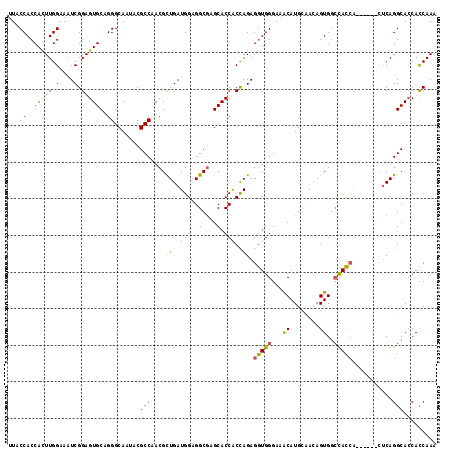
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:19 2006