
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,006,888 – 15,006,979 |
| Length | 91 |
| Max. P | 0.911970 |

| Location | 15,006,888 – 15,006,979 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 88.24 |
| Mean single sequence MFE | -32.95 |
| Consensus MFE | -26.06 |
| Energy contribution | -28.75 |
| Covariance contribution | 2.69 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
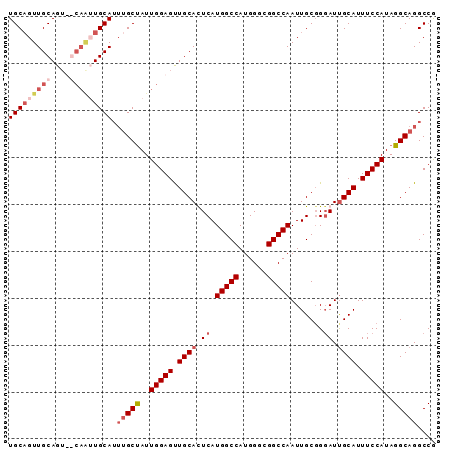
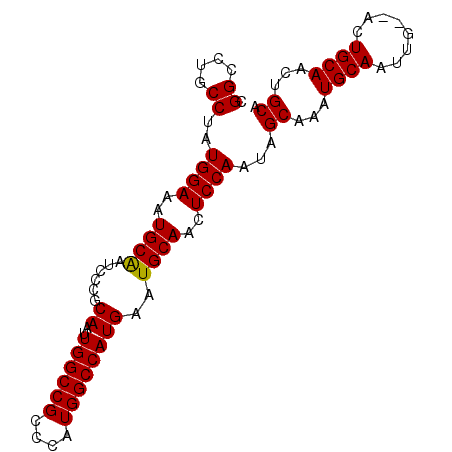
>X_DroMel_CAF1 15006888 91 + 22224390 UGCUGUUGCAGUUGCAGUUGCAUUUGCUAUUGGAGUUGCAUUCAUGGCCAUGGGCGGCCAAUUGCGGGAUUGCAUUUCCAUAGGCAGGCCA (((..((((....))))..)))((((((..(((((.((((.((.(((((......))))).......)).)))).)))))..))))))... ( -32.40) >DroSec_CAF1 24560 86 + 1 UGCAGUUGCAG---CAAAUGCA--AGCUAUUGGAGUUGCAUUCAUGGCCAUGGUCGGCCAAUUGCGGGAUCGCAUUUCCAUAGGCAGGCCG .((..(((((.---....))))--)(((..(((((.(((((((.(((((......)))))......)))).))).)))))..)))..)).. ( -28.80) >DroEre_CAF1 25619 85 + 1 UGCAGUUGCAGU------UGCAUUUGCCAUUGGAGCUGCACUCAUGGCCAUGGGCGGCCAAUUGCCGUAUUGCAUUUCCAUAGGCAGGCCG (((((((.((((------.((....)).)))).))))))).....((((....(((((.....)))))..(((..........))))))). ( -33.70) >DroYak_CAF1 24797 91 + 1 UGCAGUUGCAGUUGCAAUUGCAUUUGCUAUUGGAGUUGCACUCAUGGCCAUGGGCGGCCAAUUGCGGGAUUGCAUUUCCAUAGGCAGGCCG (((((((((....)))))))))((((((..(((((.((((.((.(((((......))))).......)).)))).)))))..))))))... ( -36.90) >consensus UGCAGUUGCAGU__CAAUUGCAUUUGCUAUUGGAGUUGCACUCAUGGCCAUGGGCGGCCAAUUGCGGGAUUGCAUUUCCAUAGGCAGGCCG (((((((((....))))))))).(((((..(((((.((((.((.(((((......))))).......)).)))).)))))..))))).... (-26.06 = -28.75 + 2.69)


| Location | 15,006,888 – 15,006,979 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 88.24 |
| Mean single sequence MFE | -26.15 |
| Consensus MFE | -23.34 |
| Energy contribution | -23.15 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
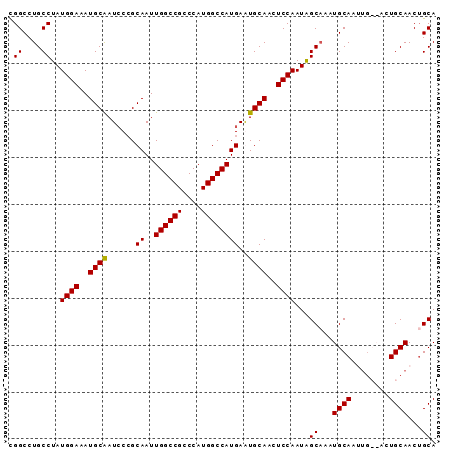
>X_DroMel_CAF1 15006888 91 - 22224390 UGGCCUGCCUAUGGAAAUGCAAUCCCGCAAUUGGCCGCCCAUGGCCAUGAAUGCAACUCCAAUAGCAAAUGCAACUGCAACUGCAACAGCA ..((.((....((((..((((.((.......((((((....)))))).)).))))..))))...(((..(((....)))..)))..)))). ( -25.10) >DroSec_CAF1 24560 86 - 1 CGGCCUGCCUAUGGAAAUGCGAUCCCGCAAUUGGCCGACCAUGGCCAUGAAUGCAACUCCAAUAGCU--UGCAUUUG---CUGCAACUGCA (((((..((...))...((((....))))...))))).....(((...((((((((((.....)).)--))))))))---))((....)). ( -23.60) >DroEre_CAF1 25619 85 - 1 CGGCCUGCCUAUGGAAAUGCAAUACGGCAAUUGGCCGCCCAUGGCCAUGAGUGCAGCUCCAAUGGCAAAUGCA------ACUGCAACUGCA ..((.((((..((((..((((....(((...(((....)))..))).....))))..))))..))))..(((.------...)))...)). ( -28.10) >DroYak_CAF1 24797 91 - 1 CGGCCUGCCUAUGGAAAUGCAAUCCCGCAAUUGGCCGCCCAUGGCCAUGAGUGCAACUCCAAUAGCAAAUGCAAUUGCAACUGCAACUGCA .....(((...((((..((((......((..((((((....))))))))..))))..))))...)))..((((.((((....)))).)))) ( -27.80) >consensus CGGCCUGCCUAUGGAAAUGCAAUCCCGCAAUUGGCCGCCCAUGGCCAUGAAUGCAACUCCAAUAGCAAAUGCAAUUG__ACUGCAACUGCA .((....))..((((..((((......((..((((((....))))))))..))))..))))...((...((((........))))...)). (-23.34 = -23.15 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:16 2006