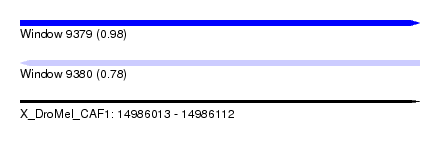
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,986,013 – 14,986,112 |
| Length | 99 |
| Max. P | 0.982506 |

| Location | 14,986,013 – 14,986,112 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 88.62 |
| Mean single sequence MFE | -23.75 |
| Consensus MFE | -20.39 |
| Energy contribution | -20.50 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
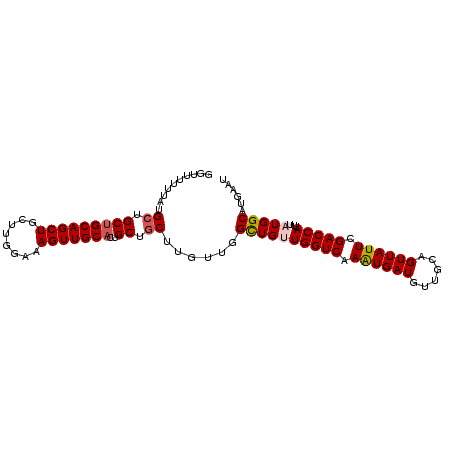
>X_DroMel_CAF1 14986013 99 + 22224390 AUUCAUGCCAUAAAUGGUCGAAUAACUGCAACAUCAUUUGACCAACAGCCAACAAGCAGCAGUGCAACUUUCCAAGCAGCUGCAGCAGCAUAAAAAACC ....((((......(((((((((............)))))))))...........((.(((((((..........))).)))).)).))))........ ( -23.10) >DroSec_CAF1 3991 99 + 1 AUUCAUGCCAUAAAUGGUCGAAUAACUGCAACAUCAUUUGACCAACAGCCAACAAGCAGCAGUGCAACUUUCCAAGCAGCUGCAGCAGCAUAAAAAACC ....((((......(((((((((............)))))))))...........((.(((((((..........))).)))).)).))))........ ( -23.10) >DroSim_CAF1 4016 99 + 1 AUUCAUGCCAUAAAUGGUCGAAUAACUGCAACAUCAUUUGACCAACAGCGAACAAGCAGCAGUGCAACUUUCCAAGCAGCUGCAGCAGCAUAAAAAACC ....((((......(((((((((............)))))))))...........((.(((((((..........))).)))).)).))))........ ( -23.10) >DroEre_CAF1 3937 99 + 1 AUUCAUGCCAUAAAUGGUCGAAUAACUGCAACAUCAUUUGACCAACAGCCAACAAGCAGCAGUGCAACUUUCCAAGCAGCUGCAGCGGCGUUAAAAAAC ....(((((.....(((((((((............)))))))))...........((.(((((((..........))).)))).)))))))........ ( -26.40) >DroYak_CAF1 3932 90 + 1 AUUCAUGCCAUAAAUGGUCGAAUAACUGCAACAUCAUUUGACCAGCAGCCAACAAGCAGCAGUGCAACUUUCCAAGCAGCUGCAGCAGCA--------- .....(((......(((((((((............)))))))))((.((......)).(((((((..........))).)))).)).)))--------- ( -22.50) >DroAna_CAF1 3222 85 + 1 AGGCAUGCCAUAAAUGGUCGAAUAACUGCAACAUCACUUGACCAGCAACAG---UGCAGCA--GCAACUUUCCAAGCAGCUGCAGCGCCA--------- .(((.(((......(((((((................))))))))))....---((((((.--((..........)).))))))..))).--------- ( -24.29) >consensus AUUCAUGCCAUAAAUGGUCGAAUAACUGCAACAUCAUUUGACCAACAGCCAACAAGCAGCAGUGCAACUUUCCAAGCAGCUGCAGCAGCAUAAAAAACC .....(((......(((((((((............)))))))))...........(((((..(((..........)))))))).)))............ (-20.39 = -20.50 + 0.11)



| Location | 14,986,013 – 14,986,112 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 88.62 |
| Mean single sequence MFE | -29.03 |
| Consensus MFE | -24.88 |
| Energy contribution | -25.27 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14986013 99 - 22224390 GGUUUUUUAUGCUGCUGCAGCUGCUUGGAAAGUUGCACUGCUGCUUGUUGGCUGUUGGUCAAAUGAUGUUGCAGUUAUUCGACCAUUUAUGGCAUGAAU .....(((((((((((((((((........)))))))..)).(((....)))...(((((.((((((......)))))).))))).....)))))))). ( -29.70) >DroSec_CAF1 3991 99 - 1 GGUUUUUUAUGCUGCUGCAGCUGCUUGGAAAGUUGCACUGCUGCUUGUUGGCUGUUGGUCAAAUGAUGUUGCAGUUAUUCGACCAUUUAUGGCAUGAAU .....(((((((((((((((((........)))))))..)).(((....)))...(((((.((((((......)))))).))))).....)))))))). ( -29.70) >DroSim_CAF1 4016 99 - 1 GGUUUUUUAUGCUGCUGCAGCUGCUUGGAAAGUUGCACUGCUGCUUGUUCGCUGUUGGUCAAAUGAUGUUGCAGUUAUUCGACCAUUUAUGGCAUGAAU .....((((((((((.((((.(((..........))))))).))...........(((((.((((((......)))))).))))).....)))))))). ( -29.50) >DroEre_CAF1 3937 99 - 1 GUUUUUUAACGCCGCUGCAGCUGCUUGGAAAGUUGCACUGCUGCUUGUUGGCUGUUGGUCAAAUGAUGUUGCAGUUAUUCGACCAUUUAUGGCAUGAAU ..........((((((((((((........)))))))..)).(((....)))...(((((.((((((......)))))).))))).....)))...... ( -27.80) >DroYak_CAF1 3932 90 - 1 ---------UGCUGCUGCAGCUGCUUGGAAAGUUGCACUGCUGCUUGUUGGCUGCUGGUCAAAUGAUGUUGCAGUUAUUCGACCAUUUAUGGCAUGAAU ---------((((..(((((((........)))))))..((.(((....))).))(((((.((((((......)))))).))))).....))))..... ( -26.90) >DroAna_CAF1 3222 85 - 1 ---------UGGCGCUGCAGCUGCUUGGAAAGUUGC--UGCUGCA---CUGUUGCUGGUCAAGUGAUGUUGCAGUUAUUCGACCAUUUAUGGCAUGCCU ---------.(((((.(((((.(((.....))).))--))).)).---((((...(((((.((((((......)))))).)))))...))))...))). ( -30.60) >consensus GGUUUUUUAUGCUGCUGCAGCUGCUUGGAAAGUUGCACUGCUGCUUGUUGGCUGUUGGUCAAAUGAUGUUGCAGUUAUUCGACCAUUUAUGGCAUGAAU ..........((.(((((((((........)))))))..)).))......((((((((((.((((((......)))))).)))))...)))))...... (-24.88 = -25.27 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:13 2006