
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,984,930 – 14,985,020 |
| Length | 90 |
| Max. P | 0.976497 |

| Location | 14,984,930 – 14,985,020 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 90.38 |
| Mean single sequence MFE | -21.02 |
| Consensus MFE | -16.44 |
| Energy contribution | -16.20 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
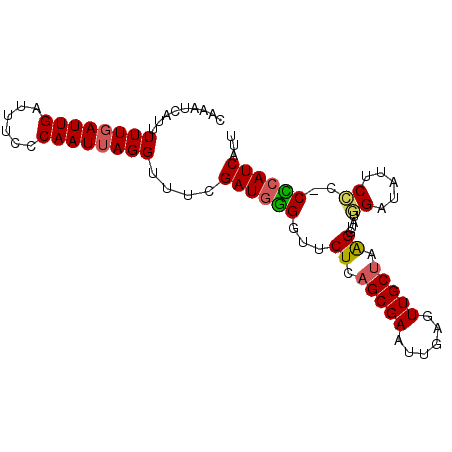
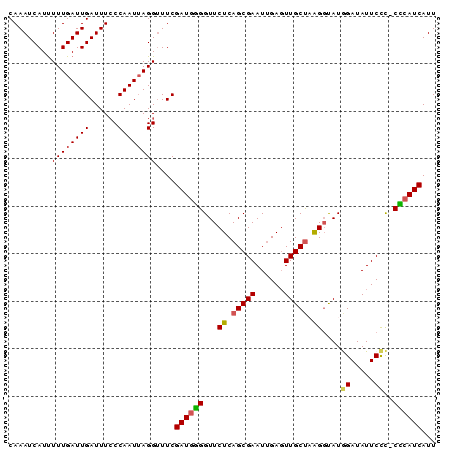
>X_DroMel_CAF1 14984930 90 + 22224390 CAAAUCAUUUUUGAUUGAUUUCCCAAUUAGGUUUCGAUGGGGUUCUCAGCGAAUUGAGUUGCUAAGGUAUGGAUAUUCCCCCCCAUCAUU .........((((((((......))))))))....(((((((.(((.(((((......))))).)))...((.....)).)))))))... ( -23.60) >DroSec_CAF1 2875 89 + 1 CAAAUCAUUUUUGAUUGAUUUCCCAAUAAGGUUUCGAUGCGGUUCUCAGCGAAUAGAUUUGCUAAGGUAUGGAUAUUCCC-CGCAUCAUU ..(((((....)))))((...((......))..))(((((((.(((.((((((....)))))).)))...((.....)))-))))))... ( -23.50) >DroSim_CAF1 2910 89 + 1 CAAAUCAUUUUUGAUUGAUUUCCCAAUUAGGUUUCGAUGGGGUUCUCAGCGAAUUGAGUUGCUAAGGCAUGGAUAUUCCC-CCGAUCAUU .........((((((((......))))))))....(((.(((.(((.(((((......))))).)))...((.....)))-)).)))... ( -19.10) >DroEre_CAF1 2889 89 + 1 CAAAUCAUUUUUGAUUGAUUUCCCAAUUAGGCUUCGAUGGGGUUCUCAGCGAAUUGAGUUGCCUGGGCAUAGAUAUUCUC-CCCAUCAUU .........((((((((......))))))))....(((((((..(((((....))))).(((....)))..........)-))))))... ( -22.90) >DroYak_CAF1 2842 89 + 1 CAAAUCAUCUUUGAUUGAUUUACCAAUUAGGUUUCGAUGUGGUUCUCAGCGAAUUGAGUUGCUUGGCUAUAGAUAUUCCU-CACAUCAUU ..(((((....)))))((...(((.....))).))((((((...((.(((((......))))).))....((......))-))))))... ( -16.00) >consensus CAAAUCAUUUUUGAUUGAUUUCCCAAUUAGGUUUCGAUGGGGUUCUCAGCGAAUUGAGUUGCUAAGGUAUGGAUAUUCCC_CCCAUCAUU .........((((((((......))))))))....((((((...((.(((((......))))).))....((.....))..))))))... (-16.44 = -16.20 + -0.24)

| Location | 14,984,930 – 14,985,020 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 90.38 |
| Mean single sequence MFE | -23.32 |
| Consensus MFE | -17.62 |
| Energy contribution | -17.50 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
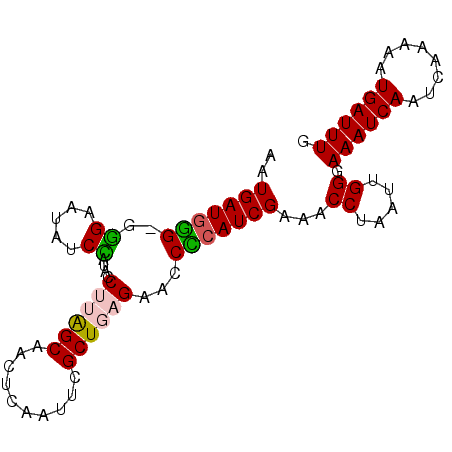
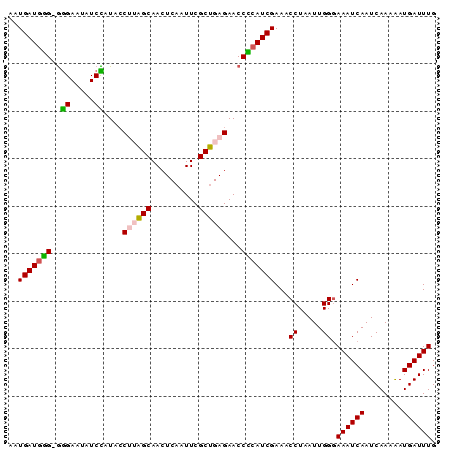
>X_DroMel_CAF1 14984930 90 - 22224390 AAUGAUGGGGGGGAAUAUCCAUACCUUAGCAACUCAAUUCGCUGAGAACCCCAUCGAAACCUAAUUGGGAAAUCAAUCAAAAAUGAUUUG ..((((((((.((.....))....((((((..........))))))..))))))))...(((....)))((((((........)))))). ( -27.10) >DroSec_CAF1 2875 89 - 1 AAUGAUGCG-GGGAAUAUCCAUACCUUAGCAAAUCUAUUCGCUGAGAACCGCAUCGAAACCUUAUUGGGAAAUCAAUCAAAAAUGAUUUG ..(((((((-(((.....))....((((((..........))))))..))))))))...(((....)))((((((........)))))). ( -26.60) >DroSim_CAF1 2910 89 - 1 AAUGAUCGG-GGGAAUAUCCAUGCCUUAGCAACUCAAUUCGCUGAGAACCCCAUCGAAACCUAAUUGGGAAAUCAAUCAAAAAUGAUUUG ..((((.((-(((.....))....((((((..........))))))..))).))))...(((....)))((((((........)))))). ( -20.60) >DroEre_CAF1 2889 89 - 1 AAUGAUGGG-GAGAAUAUCUAUGCCCAGGCAACUCAAUUCGCUGAGAACCCCAUCGAAGCCUAAUUGGGAAAUCAAUCAAAAAUGAUUUG ..(((((((-(..........(((....))).((((......))))..))))))))...(((....)))((((((........)))))). ( -25.50) >DroYak_CAF1 2842 89 - 1 AAUGAUGUG-AGGAAUAUCUAUAGCCAAGCAACUCAAUUCGCUGAGAACCACAUCGAAACCUAAUUGGUAAAUCAAUCAAAGAUGAUUUG ..(((((((-.((...........))......((((......))))...)))))))..(((.....)))((((((........)))))). ( -16.80) >consensus AAUGAUGGG_GGGAAUAUCCAUACCUUAGCAACUCAAUUCGCUGAGAACCCCAUCGAAACCUAAUUGGGAAAUCAAUCAAAAAUGAUUUG ..(((((((..((.....))....((((((..........))))))...)))))))...((.....)).((((((........)))))). (-17.62 = -17.50 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:11 2006