| Sequence ID | X_DroMel_CAF1 |
|---|---|
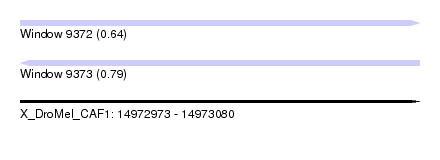
| Location | 14,972,973 – 14,973,080 |
| Length | 107 |
| Max. P | 0.793827 |

| Location | 14,972,973 – 14,973,080 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -25.72 |
| Energy contribution | -26.30 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
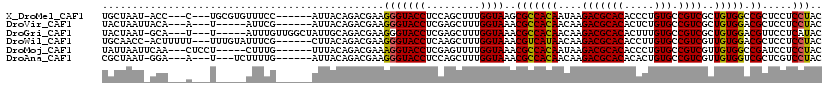
>X_DroMel_CAF1 14972973 107 + 22224390 GUAGGAGGAGCGGCCACAGCGACGGCACAGGGUGUGCGUCUUAUUGUGGCGCUUACCAAAGCUGGAGGUACCCUUCGUCUGUAAU------GGAAACACGCA---G---GGU-AUUAGCA ...((..(((((.((((((.((((.(((.....)))))))...))))))))))).))...((((...(((((((.(((.......------(....)))).)---)---)))-)))))). ( -41.61) >DroVir_CAF1 276 103 + 1 GUAGGAGGAGCGUCCACAGCGACGGCACAGAGUGUGCGUCUUGUUGUGGCGUUUACCAAAGCUCGAGGUACCCUUCGUCUGUAAU------CGAAU-----A---U---UGUAAUUAGUA ...(((((((((.(((((((((((.(((.....)))))))..)))))))))))((((.........)))).))))).........------.....-----.---.---........... ( -31.20) >DroGri_CAF1 264 108 + 1 GUAUGAGGAACGUCCACAGCGACGGCACAAAGUGUGCGUCUUGUUGUGGCGUUUACCAAAGCUCGAGGUACCCUUCGUCUGCAAUAGCCAACAAAU-----A---A---UGC-AUUAGUA (((((..(((((.(((((((((((.(((.....)))))))..))))))))))))..)...((.(((((....)))))...))..............-----.---)---)))-....... ( -33.50) >DroWil_CAF1 2328 110 + 1 GUAGGAGGAGCGUCCACAACGACGGCACAAGGUGUGCGUCUUGUUAUGACGUUUACCAAAGCUUGAGGUACCCUUCGUCUGUAAG------CGAAAUACAAA---AAAAAGU-GGUUGCA .........(((.((((...((((.(((.....)))))))((((.((..(((((((...((..(((((....))))).)))))))------))..)))))).---.....))-)).))). ( -30.30) >DroMoj_CAF1 273 106 + 1 GUAGGAGGAUCGGCCACAACGACGGCACAGGGUGUGCGUCUUAUUGUGGCGUUUACCAAAACUCGAGGUACCUUUCGUCUGUAAA------CAAAG-----AGGAG---UUGAAUUAAUA ....(((((((.(((((((.((((.(((.....)))))))...)))))))((((....))))....))).))))((.(((.....------...))-----).)).---........... ( -29.50) >DroAna_CAF1 272 104 + 1 GUAGGACGAGCGACCACAACGACGGCACAGUGUGUGCGUCUUGUUGUGGCGUUUACCAAAGCUGGAGGUACCCUUCGUCUGUAAU------CAAAAGA---A---U---UCC-AUUAGCG ...((..(((((.(((((((((((.(((.....)))))))..)))))))))))).))...((((((((...))))).(((.....------....)))---.---.---...-...))). ( -33.10) >consensus GUAGGAGGAGCGUCCACAACGACGGCACAGGGUGUGCGUCUUGUUGUGGCGUUUACCAAAGCUCGAGGUACCCUUCGUCUGUAAU______CAAAA_____A___A___UGU_AUUAGCA ...(((((((((.(((((((((((.(((.....)))))))..)))))))))))((((.........)))).)))))............................................ (-25.72 = -26.30 + 0.58)

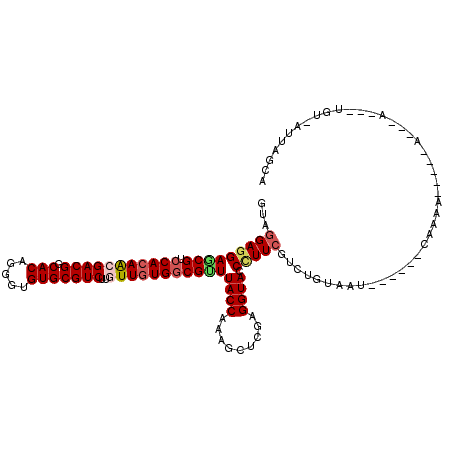
| Location | 14,972,973 – 14,973,080 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -20.68 |
| Energy contribution | -20.43 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14972973 107 - 22224390 UGCUAAU-ACC---C---UGCGUGUUUCC------AUUACAGACGAAGGGUACCUCCAGCUUUGGUAAGCGCCACAAUAAGACGCACACCCUGUGCCGUCGCUGUGGCCGCUCCUCCUAC .(((..(-(((---(---(...(((((..------.....))))).)))))).....)))...((..(((((((((....(((((((.....))).))))..))))).))))...))... ( -35.50) >DroVir_CAF1 276 103 - 1 UACUAAUUACA---A---U-----AUUCG------AUUACAGACGAAGGGUACCUCGAGCUUUGGUAAACGCCACAACAAGACGCACACUCUGUGCCGUCGCUGUGGACGCUCCUCCUAC ...........---.---.-----.((((------........))))(((((((.........))))..(((((((....(((((((.....))).))))..))))).)).....))).. ( -23.80) >DroGri_CAF1 264 108 - 1 UACUAAU-GCA---U---U-----AUUUGUUGGCUAUUGCAGACGAAGGGUACCUCGAGCUUUGGUAAACGCCACAACAAGACGCACACUUUGUGCCGUCGCUGUGGACGUUCCUCAUAC ((((((.-((.---.---.-----.((((((.((....)).))))))(((...)))..)).))))))(((((((((....(((((((.....))).))))..))))).))))........ ( -32.80) >DroWil_CAF1 2328 110 - 1 UGCAACC-ACUUUUU---UUUGUAUUUCG------CUUACAGACGAAGGGUACCUCAAGCUUUGGUAAACGUCAUAACAAGACGCACACCUUGUGCCGUCGUUGUGGACGCUCCUCCUAC .((....-.(((((.---((((((.....------..)))))).))))).((((.........))))...(((...(((((((((((.....))).)))).)))).)))))......... ( -28.20) >DroMoj_CAF1 273 106 - 1 UAUUAAUUCAA---CUCCU-----CUUUG------UUUACAGACGAAAGGUACCUCGAGUUUUGGUAAACGCCACAAUAAGACGCACACCCUGUGCCGUCGUUGUGGCCGAUCCUCCUAC ...........---..(((-----.((((------((....)))))))))......(((..((((....)((((((((..(((((((.....))).)))))))))))))))..))).... ( -29.10) >DroAna_CAF1 272 104 - 1 CGCUAAU-GGA---A---U---UCUUUUG------AUUACAGACGAAGGGUACCUCCAGCUUUGGUAAACGCCACAACAAGACGCACACACUGUGCCGUCGUUGUGGUCGCUCGUCCUAC .(.((((-.((---(---.---...))).------))))).(((((.(..((((.........))))..)((((((((..(((((((.....))).))))))))))))...))))).... ( -32.90) >consensus UACUAAU_ACA___A___U_____UUUCG______AUUACAGACGAAGGGUACCUCGAGCUUUGGUAAACGCCACAACAAGACGCACACCCUGUGCCGUCGCUGUGGACGCUCCUCCUAC ...............................................(((((((.........))))..(((((((....(((((((.....))).))))..))))).)).....))).. (-20.68 = -20.43 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:07 2006