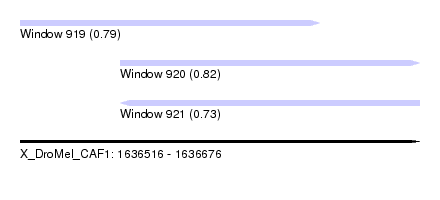
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,636,516 – 1,636,676 |
| Length | 160 |
| Max. P | 0.815440 |

| Location | 1,636,516 – 1,636,636 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.00 |
| Mean single sequence MFE | -34.92 |
| Consensus MFE | -22.53 |
| Energy contribution | -23.23 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.790391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1636516 120 + 22224390 CAGCUAAGGCGGCACUUGCUUCGUUAUGCAAUAUUUCCUACAGUCCAAUGGUGGUGCCACAGAAGAACGUACCCCUGCCAAUCGACGACAAGUCGUCAUCGAUGGAGUUGCCUCAGAUAC ...((.(((((((..((((........))))............((((.(((((((((...........)))))...))))...(((((....))))).....))))))))))).)).... ( -34.90) >DroVir_CAF1 2450 120 + 1 CCGCUAAAGCCGCGCUUGCAUCUUUGUGCAAUAUUUCUUAUAGUCCGAUGAUGGCGCCACAGAAGAAUGCGCCAUUACCAAUAGAUGAUAAGUCAUCAUCAAUGGAAUUACCGCAAAUAC ..((....)).(((.((((((....))))))............((((.(((((((((...........))))))))).)....((((((.....))))))...))).....)))...... ( -31.10) >DroGri_CAF1 2038 120 + 1 CCGCCAAGGCAGCGCUUGCCUCGCUGUGCAAUAUCUCAUAUAGUCCAAUGAUGGCACCGCAGAAGAACGCACCGUUGCCCAUUGAUGACAAAUCGUCGUCAAUGGAAUUGCCGCAAAUAC ..((...(((((((..(((((((.((..(.((((...)))).)..)).))).)))).)))........(((....)))(((((((((((.....)))))))))))...))))))...... ( -38.40) >DroWil_CAF1 16986 120 + 1 CUGCCAAGGCCGCACUUGCCUCGCUGUGCAAUAUCUCGUAUAGUCCAAUGAUGGCACCCCAAAAGAAUGUGCCGCUGCCCAUUGAUGAUAAGACAUCGUCAAUGGAAUUGCCACAAAUCC .(((((((((.......)))).(((((((........))))))).......)))))...........((((..((...(((((((((((.....)))))))))))....))))))..... ( -35.40) >DroMoj_CAF1 3328 120 + 1 CCGCGAAGGCUGCUUUAGCCUCUCUGUGCAAUAUAUCUUACAGUCCGAUGAUGGCACCACAGAAAAAUGCGCCGUUACCCAUUGACGAGAAGUCAUCGUCGAUGGAAUUACCACAAAUAC .((((.((((((...)))))).((((((.....((((............))))....))))))....)))).......((((((((((.......))))))))))............... ( -33.30) >DroAna_CAF1 1301 120 + 1 CUGCCAAAGCGGCUCUGGCCUCGCUCUGCAACAUCUCGUAUAGCCCGAUGGUGGUGCCACAAAAGAACGUGCCACUGCCCAUCGACGACAAGUCAUCGUCCAUGGAACUGCCCCAGAUCC ..(((.....)))(((((....(((.(((........))).)))(((((((..(((.(((........))).)))..).))))(((((.......)))))...)).......)))))... ( -36.40) >consensus CCGCCAAGGCCGCACUUGCCUCGCUGUGCAAUAUCUCGUAUAGUCCAAUGAUGGCACCACAGAAGAACGCGCCGCUGCCCAUUGACGACAAGUCAUCGUCAAUGGAAUUGCCACAAAUAC ..((....)).((....))........(((((................(((((((((...........))))))))).(((((((((((.....))))))))))).)))))......... (-22.53 = -23.23 + 0.70)
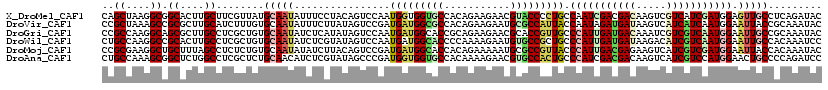
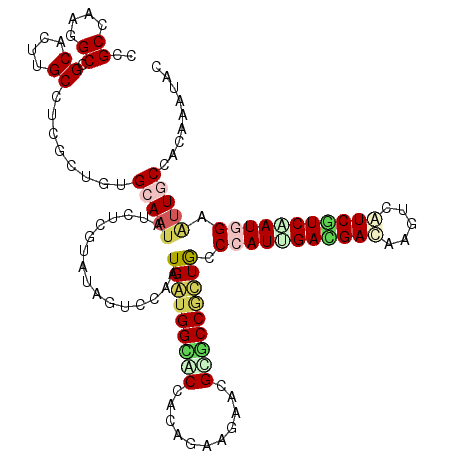
| Location | 1,636,556 – 1,636,676 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -36.88 |
| Consensus MFE | -23.68 |
| Energy contribution | -23.93 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1636556 120 + 22224390 CAGUCCAAUGGUGGUGCCACAGAAGAACGUACCCCUGCCAAUCGACGACAAGUCGUCAUCGAUGGAGUUGCCUCAGAUACACGCGGAUACGAUUGGUCGGUUGGUCUUAGAAAAGUUCAU .((.(((((((.(((((...........))))))).((((((((((((....)))))(((...((.....))...)))............)))))))..))))).))............. ( -33.30) >DroVir_CAF1 2490 120 + 1 UAGUCCGAUGAUGGCGCCACAGAAGAAUGCGCCAUUACCAAUAGAUGAUAAGUCAUCAUCAAUGGAAUUACCGCAAAUACAUGCGGACACUAUCGGACGGUUAGUGUUGGAAAAGUUCAU ..(((((((((((((((...........)))))))).(((...((((((.....))))))..))).....(((((......))))).....)))))))...................... ( -38.20) >DroGri_CAF1 2078 120 + 1 UAGUCCAAUGAUGGCACCGCAGAAGAACGCACCGUUGCCCAUUGAUGACAAAUCGUCGUCAAUGGAAUUGCCGCAAAUACACGCAGAUACUAUCGGACGCUUAGUGUUGGAGAAAUUCAU ..((((......((((..((((..(......)..))))(((((((((((.....)))))))))))...))))((........))..........)))).........((((....)))). ( -36.10) >DroWil_CAF1 17026 120 + 1 UAGUCCAAUGAUGGCACCCCAAAAGAAUGUGCCGCUGCCCAUUGAUGAUAAGACAUCGUCAAUGGAAUUGCCACAAAUCCAUGCGGACACAAUUGGUCGCCUGGUAUUGGAGAAAUUCAU ...(((((((..(((...((((.....((((((((...(((((((((((.....)))))))))))..(((...)))......)))).)))).))))..)))...)))))))......... ( -39.30) >DroMoj_CAF1 3368 120 + 1 CAGUCCGAUGAUGGCACCACAGAAAAAUGCGCCGUUACCCAUUGACGAGAAGUCAUCGUCGAUGGAAUUACCACAAAUACAUGCGGAUACCAUUGGACGGUUGGUAUUGGAAAAAUUCAU ..(((((((((((((.(...........).)))))...((((((((((.......)))))))))).....((.((......)).))....))))))))...................... ( -33.70) >DroAna_CAF1 1341 120 + 1 UAGCCCGAUGGUGGUGCCACAAAAGAACGUGCCACUGCCCAUCGACGACAAGUCAUCGUCCAUGGAACUGCCCCAGAUCCAUGCGGACACCAUUGGCCGUCUGGUGUUGGAGAAGUUCAU ......(((((..(((.(((........))).)))..).))))(((((.......)))))....(((((.(.((((..(((.((((.((....)).)))).)))..)))).).))))).. ( -40.70) >consensus UAGUCCAAUGAUGGCACCACAGAAGAACGCGCCGCUGCCCAUUGACGACAAGUCAUCGUCAAUGGAAUUGCCACAAAUACAUGCGGACACCAUUGGACGGUUGGUGUUGGAAAAAUUCAU ..(.(((((((((((((...........))))))))..(((((((((((.....)))))))))))..........................))))).)...................... (-23.68 = -23.93 + 0.26)

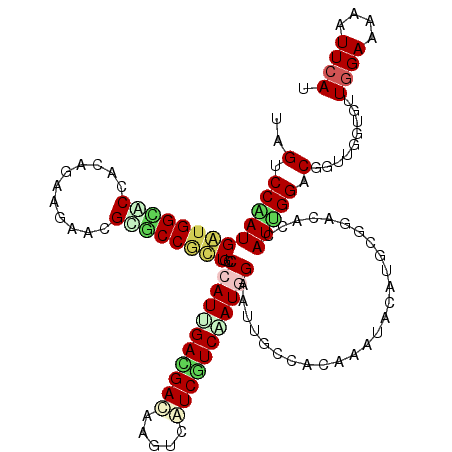
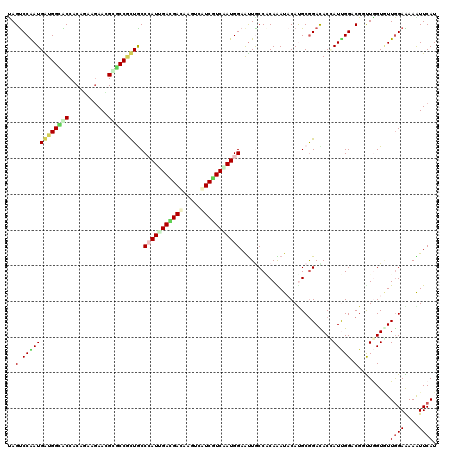
| Location | 1,636,556 – 1,636,676 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -37.32 |
| Consensus MFE | -23.65 |
| Energy contribution | -22.85 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.59 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1636556 120 - 22224390 AUGAACUUUUCUAAGACCAACCGACCAAUCGUAUCCGCGUGUAUCUGAGGCAACUCCAUCGAUGACGACUUGUCGUCGAUUGGCAGGGGUACGUUCUUCUGUGGCACCACCAUUGGACUG ........................((((((((((((.(.(((....(((....))).(((((((((.....)))))))))..))))))))))).......(((....))).))))).... ( -33.60) >DroVir_CAF1 2490 120 - 1 AUGAACUUUUCCAACACUAACCGUCCGAUAGUGUCCGCAUGUAUUUGCGGUAAUUCCAUUGAUGAUGACUUAUCAUCUAUUGGUAAUGGCGCAUUCUUCUGUGGCGCCAUCAUCGGACUA ......................(((((((.....(((((......))))).....(((..(((((((...)))))))...)))..(((((((...(....)..))))))).))))))).. ( -38.30) >DroGri_CAF1 2078 120 - 1 AUGAAUUUCUCCAACACUAAGCGUCCGAUAGUAUCUGCGUGUAUUUGCGGCAAUUCCAUUGACGACGAUUUGUCAUCAAUGGGCAACGGUGCGUUCUUCUGCGGUGCCAUCAUUGGACUA ......................(((((((.......(((......)))((((...(((((((.(((.....))).)))))))(((..((......))..)))..))))...))))))).. ( -33.10) >DroWil_CAF1 17026 120 - 1 AUGAAUUUCUCCAAUACCAGGCGACCAAUUGUGUCCGCAUGGAUUUGUGGCAAUUCCAUUGACGAUGUCUUAUCAUCAAUGGGCAGCGGCACAUUCUUUUGGGGUGCCAUCAUUGGACUA .........((((((....((((.((((.((((((((((......))))((...((((((((.((((...)))).))))))))..)))))))).....))))..))))...))))))... ( -38.60) >DroMoj_CAF1 3368 120 - 1 AUGAAUUUUUCCAAUACCAACCGUCCAAUGGUAUCCGCAUGUAUUUGUGGUAAUUCCAUCGACGAUGACUUCUCGUCAAUGGGUAACGGCGCAUUUUUCUGUGGUGCCAUCAUCGGACUG ......................((((.(((((..(((((......)))))....(((((.(((((.(....)))))).)))))....(((((((......)).)))))))))).)))).. ( -34.70) >DroAna_CAF1 1341 120 - 1 AUGAACUUCUCCAACACCAGACGGCCAAUGGUGUCCGCAUGGAUCUGGGGCAGUUCCAUGGACGAUGACUUGUCGUCGAUGGGCAGUGGCACGUUCUUUUGUGGCACCACCAUCGGGCUA ..(((((.(((((...(((..(((.((....)).)))..)))...))))).)))))....((((((.....))))))(((((...(((.((((......)))).)))..)))))...... ( -45.60) >consensus AUGAACUUCUCCAACACCAACCGUCCAAUAGUAUCCGCAUGUAUUUGCGGCAAUUCCAUUGACGAUGACUUGUCAUCAAUGGGCAACGGCACAUUCUUCUGUGGCGCCAUCAUCGGACUA ........................(((((.(((.(((((......((((.(...(((((.((((((.....)))))).)))))....).))))......))))))))....))))).... (-23.65 = -22.85 + -0.80)

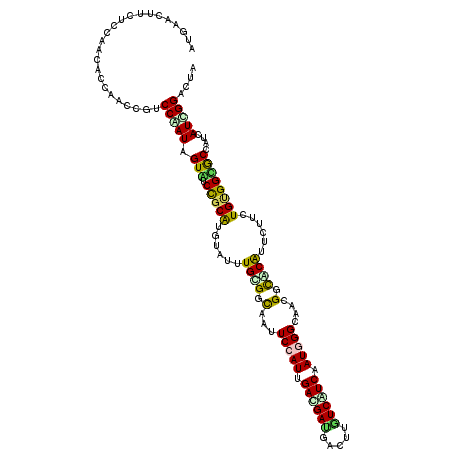
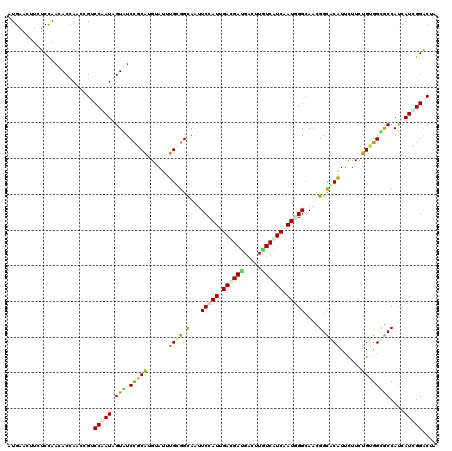
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:22 2006