
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,954,453 – 14,954,594 |
| Length | 141 |
| Max. P | 0.918822 |

| Location | 14,954,453 – 14,954,555 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.19 |
| Mean single sequence MFE | -28.70 |
| Consensus MFE | -5.09 |
| Energy contribution | -4.17 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.18 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
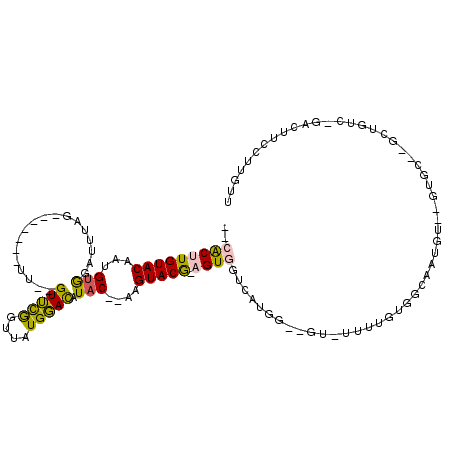
>X_DroMel_CAF1 14954453 102 + 22224390 -UGGCUGGCUGCUGUUAGUUGUUCGUUUUUAGCUAUU-----CACUUGUACAAUGUGGAUUUAG-------UU--GUUCGGUUAUGGACAUAC--AAGUACG-AGUGGUCAUGGCAGUAU -......((((((((((((((........)))))).(-----(((((((((..((((.......-------.(--(((((....)))))))))--).)))))-)))))..)))))))).. ( -29.30) >DroPse_CAF1 4655 113 + 1 GUGGUUCGUUGUUGUUUGGUGUUCAUUAUUUGUUCGUUUUAGAGCUUGUACAAUGUAUAGUAUGUAUGUACAUGAGUACACUUAUGUAUGUACAUAUGUACAAUGUGCGCAUA------- ..................((((.........((((......))))..(((((.((((((.((((((((((((((((....)))))))))))))))))))))).))))))))).------- ( -36.40) >DroSim_CAF1 891 102 + 1 -UGGUUGGCUGCUGUUAGUUGUUCGUUUUUAGCUAUU-----CACUUGUACAAUGUGGAUUUAG-------UU--GUUUGGUUAUGGACAUAC--AAGUACG-AGUGGUCAUGGAUGUGU -....((((..((..((.((((..(((..((((((..-----((((....(.....).....))-------.)--)..))))))..)))..))--)).))..-))..))))......... ( -23.20) >DroYak_CAF1 874 103 + 1 -UGGUUGGCUGCUGUUAGUUGUUCGUUUUUAGCUAUU-----CACUUGUACAUUGUGGAUUUAG-------UU--GUUUGGUUAUGGACAUAC--AAGUACGGAGUGGUCAUGGAGGUAU -....((((..((.(((.((((..(((..((((((..-----((((....(.....).....))-------.)--)..))))))..)))..))--)).)).).))..))))......... ( -22.80) >DroPer_CAF1 4626 113 + 1 GUGGUUCGUUGUUGUUUGGUGUUCAUUAUUUGUUCGUUUUAGAGCUUGUACAAUGUAUAGUAUGUAUGUACAUGAGUACACUUAUGUAUGUCCAUAUGUACAAUGUGCGCAUA------- ..................((((.........((((......))))..(((((.((((((.((((.(((((((((((....))))))))))).)))))))))).))))))))).------- ( -31.80) >consensus _UGGUUGGCUGCUGUUAGUUGUUCGUUUUUAGCUAUU_____CACUUGUACAAUGUGGAUUUAG_______UU__GUUCGGUUAUGGACAUAC__AAGUACG_AGUGGUCAUGG__GU_U ..........................................(((((((((...(((..................(((((....))))).)))....))))).))))............. ( -5.09 = -4.17 + -0.92)



| Location | 14,954,489 – 14,954,594 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.23 |
| Mean single sequence MFE | -29.06 |
| Consensus MFE | -5.09 |
| Energy contribution | -4.17 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.18 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14954489 105 + 22224390 --CACUUGUACAAUGUGGAUUUAG-------UU--GUUCGGUUAUGGACAUAC--AAGUACG-AGUGGUCAUGGCAGUAUUUUGUGGCAAUGUGUGUGCCCGCUGUU-GACUUCCUUGUU --(((((((((..((((.......-------.(--(((((....)))))))))--).)))))-))))((((..(((((.....(.((((.......)))))))))))-)))......... ( -27.80) >DroPse_CAF1 4695 105 + 1 AGAGCUUGUACAAUGUAUAGUAUGUAUGUACAUGAGUACACUUAUGUAUGUACAUAUGUACAAUGUGCGCAUA-------UUUCUGGUAAAGC--GU-----CUUUC-GACUUCCUCGUU .(((((((((((.((((((.((((((((((((((((....)))))))))))))))))))))).))))).((..-------....))...))))--((-----(....-)))....))... ( -35.70) >DroSim_CAF1 927 103 + 1 --CACUUGUACAAUGUGGAUUUAG-------UU--GUUUGGUUAUGGACAUAC--AAGUACG-AGUGGUCAUGGAUGUGUUUUGUGGCAAUGU--GUGCCCGCUGUC-GACUUCCUUGUU --(((((((((..((((..((((.-------..--.........))))..)))--).)))))-))))..((.(((.((.....((((((....--.)).))))....-.)).))).)).. ( -23.70) >DroYak_CAF1 910 105 + 1 --CACUUGUACAUUGUGGAUUUAG-------UU--GUUUGGUUAUGGACAUAC--AAGUACGGAGUGGUCAUGGAGGUAUUUUGUGGCAAUGU--GUGCUUGAUGUCGGACUUCCUUGUU --.......(((....(((.....-------..--....((((...(((((.(--(((((((.(...((((..(((...)))..))))..).)--)))))))))))).))))))).))). ( -27.00) >DroPer_CAF1 4666 105 + 1 AGAGCUUGUACAAUGUAUAGUAUGUAUGUACAUGAGUACACUUAUGUAUGUCCAUAUGUACAAUGUGCGCAUA-------UUUCUGGUAAAGC--GU-----CUUUC-GACUUCCUCGUU .(((((((((((.((((((.((((.(((((((((((....))))))))))).)))))))))).))))).((..-------....))...))))--((-----(....-)))....))... ( -31.10) >consensus __CACUUGUACAAUGUGGAUUUAG_______UU__GUUCGGUUAUGGACAUAC__AAGUACG_AGUGGUCAUGG__GU_UUUUGUGGCAAUGU__GUGC__GCUGUC_GACUUCCUUGUU ..(((((((((...(((..................(((((....))))).)))....))))).))))..................................................... ( -5.09 = -4.17 + -0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:52 2006