
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,937,351 – 14,937,494 |
| Length | 143 |
| Max. P | 0.997000 |

| Location | 14,937,351 – 14,937,461 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 82.83 |
| Mean single sequence MFE | -32.04 |
| Consensus MFE | -21.60 |
| Energy contribution | -25.35 |
| Covariance contribution | 3.75 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.65 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.997000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14937351 110 + 22224390 AUAAAUGAAAGACCUUGAUAUUUUUAUUUCGUUGUUUGGG-UCACCAAACAGCGAAUAGAAAAGC--AAAGGGUGUCACCUGAAUGGUGUUGAUCUGGGUCAACCAAUAUCAU ..........(((((.(((.((((((((.((((((((((.-...))))))))))))))))))...--.........((((.....))))...))).)))))............ ( -35.00) >DroSim_CAF1 2088 110 + 1 AUAAAUGAAAGACCUUGAUAUUUUUAUUUCGUUGUUUGGG-UCACCAAACAGCGAAUAGAAAAGC--AAAGGGUGUCACCUGAAUGGUGUUGAUCUGGAUCAACCAAUAUCAU ....((((...(((((....(((((..((((((((((((.-...))))))))))))..)))))..--..)))))...(((.....)))((((((....)))))).....)))) ( -34.20) >DroEre_CAF1 1828 104 + 1 AUAAAUGAGAGACCUUGAUAUUUUCAUCUUGUUGUUUGGGUUCACCAAACAGCGAAUAGAAAAGC--AAAGGGUGUCACCUGAAUGGUGUUGAUCUGGGUCAA-------CAU ........((.(((((....(((((...(((((((((((.....)))))))))))...)))))..--..))))).))(((.....)))((((((....)))))-------).. ( -37.10) >DroYak_CAF1 2142 90 + 1 AUAAAUGAGAGACCUUGAUAUUAU----------------UUCACCAAACAGCGAAGUGAAAAGGCAAAAGGGUGUCACCUCAAUGGUGUUGAUCUGGGUCAA-------CAU ........((.(((((.......(----------------(((((...........)))))).......))))).))(((.....)))((((((....)))))-------).. ( -21.84) >consensus AUAAAUGAAAGACCUUGAUAUUUUUAUUUCGUUGUUUGGG_UCACCAAACAGCGAAUAGAAAAGC__AAAGGGUGUCACCUGAAUGGUGUUGAUCUGGGUCAA_______CAU ..........(((((.(((.(((((..((((((((((((.....))))))))))))..))))).............((((.....))))...))).)))))............ (-21.60 = -25.35 + 3.75)

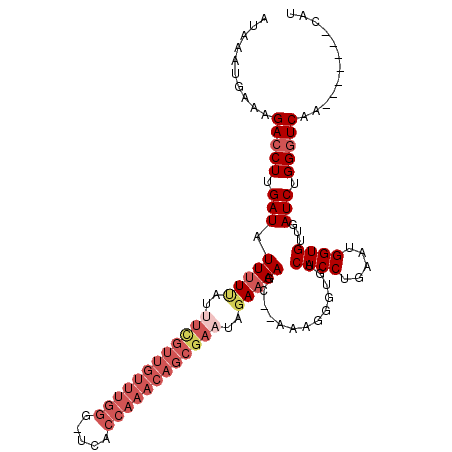

| Location | 14,937,351 – 14,937,461 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 82.83 |
| Mean single sequence MFE | -23.26 |
| Consensus MFE | -14.84 |
| Energy contribution | -14.65 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14937351 110 - 22224390 AUGAUAUUGGUUGACCCAGAUCAACACCAUUCAGGUGACACCCUUU--GCUUUUCUAUUCGCUGUUUGGUGA-CCCAAACAACGAAAUAAAAAUAUCAAGGUCUUUCAUUUAU .......(((..((((..(((...((((.....)))).........--.........((((.(((((((...-.))))))).))))........)))..))))..)))..... ( -26.40) >DroSim_CAF1 2088 110 - 1 AUGAUAUUGGUUGAUCCAGAUCAACACCAUUCAGGUGACACCCUUU--GCUUUUCUAUUCGCUGUUUGGUGA-CCCAAACAACGAAAUAAAAAUAUCAAGGUCUUUCAUUUAU .((((((((((((((....)))).((((.....))))..)))....--.........((((.(((((((...-.))))))).)))).....)))))))............... ( -25.90) >DroEre_CAF1 1828 104 - 1 AUG-------UUGACCCAGAUCAACACCAUUCAGGUGACACCCUUU--GCUUUUCUAUUCGCUGUUUGGUGAACCCAAACAACAAGAUGAAAAUAUCAAGGUCUCUCAUUUAU (((-------..((((..(((...((((.....)))).........--..(((((...((..(((((((.....)))))))....)).))))).)))..))))...))).... ( -22.40) >DroYak_CAF1 2142 90 - 1 AUG-------UUGACCCAGAUCAACACCAUUGAGGUGACACCCUUUUGCCUUUUCACUUCGCUGUUUGGUGAA----------------AUAAUAUCAAGGUCUCUCAUUUAU (((-------..((((..(((...(((((.((((((((...............)))))))).....)))))..----------------.....)))..))))...))).... ( -18.36) >consensus AUG_______UUGACCCAGAUCAACACCAUUCAGGUGACACCCUUU__GCUUUUCUAUUCGCUGUUUGGUGA_CCCAAACAACGAAAUAAAAAUAUCAAGGUCUCUCAUUUAU ............((((..(((...((((.....)))).....................(((((....)))))......................)))..)))).......... (-14.84 = -14.65 + -0.19)

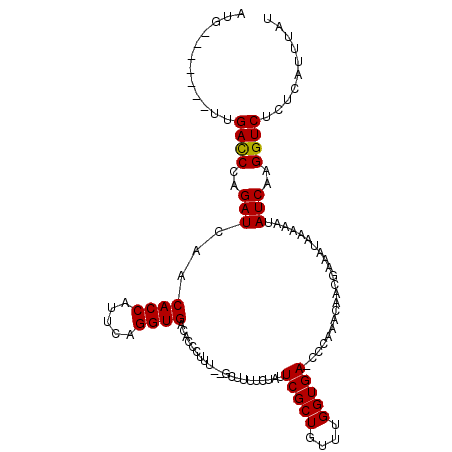
| Location | 14,937,387 – 14,937,494 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 88.37 |
| Mean single sequence MFE | -26.68 |
| Consensus MFE | -17.08 |
| Energy contribution | -17.07 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14937387 107 + 22224390 UGGG-UCACCAAACAGCGAAUAGAAAAGC--AAAGGGUGUCACCUGAAUGGUGUUGAUCUGGGUCAACCAAUAUCAUACCAAUGGAACACCAGACCCCAAAAAAUGACGG .(((-((........((..........))--....(((((..((....((((((.(((.(((.....)))..)))))))))..)).))))).)))))............. ( -29.50) >DroSim_CAF1 2124 106 + 1 UGGG-UCACCAAACAGCGAAUAGAAAAGC--AAAGGGUGUCACCUGAAUGGUGUUGAUCUGGAUCAACCAAUAUCAUACCAAUGGAACACCAGACCCCAA-AAAUGACGG .(((-((........((..........))--....(((((..((....((((((.(((.(((.....)))..)))))))))..)).))))).)))))...-......... ( -28.20) >DroEre_CAF1 1864 100 + 1 UGGGUUCACCAAACAGCGAAUAGAAAAGC--AAAGGGUGUCACCUGAAUGGUGUUGAUCUGGGUCAA-------CAUACCAAUAGAACACCAGACCCCAG-AAAUGACGG ((((.((........((..........))--....(((((...(((..((((((((((....)))))-------)..)))).))).))))).)).)))).-......... ( -25.30) >DroYak_CAF1 2166 99 + 1 ----UUCACCAAACAGCGAAGUGAAAAGGCAAAAGGGUGUCACCUCAAUGGUGUUGAUCUGGGUCAA-------CAUACCAAUGGAACACCAGACCCCAAAAAAUGACGG ----(((((...........))))).........(((.(((.......((((((((((....)))))-------)..)))).(((....)))))))))............ ( -23.70) >consensus UGGG_UCACCAAACAGCGAAUAGAAAAGC__AAAGGGUGUCACCUGAAUGGUGUUGAUCUGGGUCAA_______CAUACCAAUGGAACACCAGACCCCAA_AAAUGACGG ..................................(((.(((.......(((((((....(((................)))....)))))))))))))............ (-17.08 = -17.07 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:43 2006