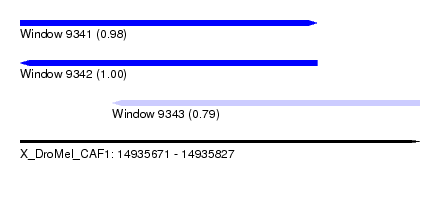
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,935,671 – 14,935,827 |
| Length | 156 |
| Max. P | 0.997640 |

| Location | 14,935,671 – 14,935,787 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.16 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -20.12 |
| Energy contribution | -20.72 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14935671 116 + 22224390 UUGAAUUUUUUGGAGAAA----AAUCAAUAUUCUGAAGUUGGCACAAAAAACAGAAACCUCUUUAAACCGCAAGAGAAAUCUGAGUGCCAUUAAAAUCUUCAUUCGGGAAAUCAUUCUAA .(((.(((((((((((..----..)).......(((((.((((((......((((...(((((........)))))...)))).)))))).......)))))))))))))))))...... ( -22.80) >DroSec_CAF1 146 115 + 1 UUGAAUUUUUUGGAGAAA----AAUGAAUAUUCUGAAGUUGGCACAA-AAACAGAAACCUCUUUAAACCGAAAGAGAAAUCUGAGUGCCAUUAAAAUCUUCAUUCGGGAAAUCAUUCUAA .(((.((((((((((((.----........)))(((((.((((((..-...((((...((((((......))))))...)))).)))))).......)))))))))))))))))...... ( -24.70) >DroSim_CAF1 411 115 + 1 UUGAAUUUUUUGGAGAAA----AAUGAAUAUUCUGAAGUUGGCACAA-AAACAGAAACCUCUUUAAACCGAAAGAGAAAUCUGAGUGCCAUUAAAAUCUUCAUUCGGGAAAUCAUUCUAA .(((.((((((((((((.----........)))(((((.((((((..-...((((...((((((......))))))...)))).)))))).......)))))))))))))))))...... ( -24.70) >DroEre_CAF1 155 114 + 1 AGGA-AUUUUUGAAGAAA----AAUGAAUAUUCUGAAGUUGGCACAG-AAACAGAAACCUCGUUAAACCGAAAGAGAAAUCUGAGUGCCAUUGAAAUCUUCAUUCGGGAAAUCAUUCUAA ....-.............----(((((...(((((((..((((((..-...........(((......))).(((....)))..)))))).(((.....))))))))))..))))).... ( -21.80) >DroYak_CAF1 438 118 + 1 UGGA-GUUUUUGAAGGAAAUGAAAUGAAUAUUCUGAAGUUGGCACAA-AAACAGAAACCUCCUUAAACCGAAAGAGAAAUCUGAGUGCCAUUAAAAUCUUCAUUCGGGAAAUCAUUAUAA ..((-.(((((((((((.((.......)).)))(((((.((((((..-...((((...(((.((......)).)))...)))).)))))).......))))))))))))).))....... ( -21.00) >consensus UUGAAUUUUUUGGAGAAA____AAUGAAUAUUCUGAAGUUGGCACAA_AAACAGAAACCUCUUUAAACCGAAAGAGAAAUCUGAGUGCCAUUAAAAUCUUCAUUCGGGAAAUCAUUCUAA ..((.((((((((((((.............)))(((((.((((((......((((...((((((......))))))...)))).)))))).......))))))))))))))))....... (-20.12 = -20.72 + 0.60)



| Location | 14,935,671 – 14,935,787 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.16 |
| Mean single sequence MFE | -26.74 |
| Consensus MFE | -22.14 |
| Energy contribution | -22.82 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.90 |
| SVM RNA-class probability | 0.997640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14935671 116 - 22224390 UUAGAAUGAUUUCCCGAAUGAAGAUUUUAAUGGCACUCAGAUUUCUCUUGCGGUUUAAAGAGGUUUCUGUUUUUUGUGCCAACUUCAGAAUAUUGAUU----UUUCUCCAAAAAAUUCAA ..((((.((((.......(((((.......((((((.((((..((((((........))))))..))))......)))))).))))).......))))----.))))............. ( -26.84) >DroSec_CAF1 146 115 - 1 UUAGAAUGAUUUCCCGAAUGAAGAUUUUAAUGGCACUCAGAUUUCUCUUUCGGUUUAAAGAGGUUUCUGUUU-UUGUGCCAACUUCAGAAUAUUCAUU----UUUCUCCAAAAAAUUCAA ...((((........(((((((.(((((..((((((.((((..(((((((......)))))))..))))...-..)))))).....))))).))))))----)...........)))).. ( -28.11) >DroSim_CAF1 411 115 - 1 UUAGAAUGAUUUCCCGAAUGAAGAUUUUAAUGGCACUCAGAUUUCUCUUUCGGUUUAAAGAGGUUUCUGUUU-UUGUGCCAACUUCAGAAUAUUCAUU----UUUCUCCAAAAAAUUCAA ...((((........(((((((.(((((..((((((.((((..(((((((......)))))))..))))...-..)))))).....))))).))))))----)...........)))).. ( -28.11) >DroEre_CAF1 155 114 - 1 UUAGAAUGAUUUCCCGAAUGAAGAUUUCAAUGGCACUCAGAUUUCUCUUUCGGUUUAACGAGGUUUCUGUUU-CUGUGCCAACUUCAGAAUAUUCAUU----UUUCUUCAAAAAU-UCCU ..(((((((.(((..((((((.....))).((((((.((((..((((............))))..))))...-..))))))..))).)))...)))))----))...........-.... ( -23.60) >DroYak_CAF1 438 118 - 1 UUAUAAUGAUUUCCCGAAUGAAGAUUUUAAUGGCACUCAGAUUUCUCUUUCGGUUUAAGGAGGUUUCUGUUU-UUGUGCCAACUUCAGAAUAUUCAUUUCAUUUCCUUCAAAAAC-UCCA ......(((......(((((((.(((((..((((((.((((..(((((((......)))))))..))))...-..)))))).....))))).)))))))........))).....-.... ( -27.04) >consensus UUAGAAUGAUUUCCCGAAUGAAGAUUUUAAUGGCACUCAGAUUUCUCUUUCGGUUUAAAGAGGUUUCUGUUU_UUGUGCCAACUUCAGAAUAUUCAUU____UUUCUCCAAAAAAUUCAA ................((((((.(((((..((((((.((((..(((((((......)))))))..))))......)))))).....))))).))))))...................... (-22.14 = -22.82 + 0.68)


| Location | 14,935,707 – 14,935,827 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.71 |
| Mean single sequence MFE | -29.16 |
| Consensus MFE | -27.72 |
| Energy contribution | -27.52 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14935707 120 - 22224390 GUAGACAACUAAUUUACUUGGGCUGGUUGUUGAGGAGGUUUUAGAAUGAUUUCCCGAAUGAAGAUUUUAAUGGCACUCAGAUUUCUCUUGCGGUUUAAAGAGGUUUCUGUUUUUUGUGCC ...((((((((.(((....))).))))))))(.(((((((.......))))))))................(((((.((((..((((((........))))))..))))......))))) ( -30.60) >DroSec_CAF1 182 119 - 1 GUAGACAACUAAUUUAUUUGGAAAGGUUGUUGCGGAGGUUUUAGAAUGAUUUCCCGAAUGAAGAUUUUAAUGGCACUCAGAUUUCUCUUUCGGUUUAAAGAGGUUUCUGUUU-UUGUGCC ((((.(((((..(((....)))..)))))))))(((((((.......))))))).................(((((.((((..(((((((......)))))))..))))...-..))))) ( -30.10) >DroSim_CAF1 447 119 - 1 GUAGACAACUAAUUUACUUGAAAAGGUUGUUGCGGAGGUUUUAGAAUGAUUUCCCGAAUGAAGAUUUUAAUGGCACUCAGAUUUCUCUUUCGGUUUAAAGAGGUUUCUGUUU-UUGUGCC ((((.(((((..(((....)))..)))))))))(((((((.......))))))).................(((((.((((..(((((((......)))))))..))))...-..))))) ( -30.50) >DroEre_CAF1 190 119 - 1 GUAGACAACUCAAUUACUUGGUAAGGUUGUUAAGGAGGUUUUAGAAUGAUUUCCCGAAUGAAGAUUUCAAUGGCACUCAGAUUUCUCUUUCGGUUUAACGAGGUUUCUGUUU-CUGUGCC ...(((((((..(((....)))..)))))))..(((((((.......))))))).(((.......)))...(((((.((((..((((............))))..))))...-..))))) ( -26.80) >DroYak_CAF1 477 119 - 1 GUAGACAACUCAAUUACUUGGUAAGGUUGCAAAGGAGAUUUUAUAAUGAUUUCCCGAAUGAAGAUUUUAAUGGCACUCAGAUUUCUCUUUCGGUUUAAGGAGGUUUCUGUUU-UUGUGCC ...(.(((((..(((....)))..))))))...(((((((.......))))))).................(((((.((((..(((((((......)))))))..))))...-..))))) ( -27.80) >consensus GUAGACAACUAAUUUACUUGGAAAGGUUGUUGAGGAGGUUUUAGAAUGAUUUCCCGAAUGAAGAUUUUAAUGGCACUCAGAUUUCUCUUUCGGUUUAAAGAGGUUUCUGUUU_UUGUGCC ...(((((((..(((....)))..)))))))..(((((((.......))))))).................(((((.((((..(((((((......)))))))..))))......))))) (-27.72 = -27.52 + -0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:40 2006