| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,920,931 – 14,921,050 |
| Length | 119 |
| Max. P | 0.858210 |

| Location | 14,920,931 – 14,921,050 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.86 |
| Mean single sequence MFE | -36.39 |
| Consensus MFE | -13.74 |
| Energy contribution | -14.38 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14920931 119 - 22224390 GAGGAAGAAGAGUCCUCCGGCUCAU-CUUCGUCUUCAUUCACCACAUCGAAAUCGAGCAGAGGCUUGCCGACCAGAGCAAUCUCAUCGAUUUUGUGGUGAAUCUUGGUUAUCUUGCCAUC ((.(((((.(((((....))))).)-)))).))...((((((((((...(((((((...((((.(((((.....).)))))))).)))))))))))))))))..((((......)))).. ( -46.70) >DroPse_CAF1 27419 115 - 1 -AGGAUUCAGUGCUAUU---CUCGU-CACCCUCCUCGUCGAGCACAUCAAAGUCCAGCAGGGGCUUGCCCACCAAUGCGAUCUCAUCGAUGCUGUGGUGGAUUUUAGUGAUUUUGCCAUC -.((.....(((((...---..((.-.........))...)))))..((((((((.((((....))))((((((..((.(((.....)))))..))))))......).)))))))))... ( -30.60) >DroSim_CAF1 19698 120 - 1 GAGGAAGAAGAGUCCUCCGCCUCAUUCUUCCUCUUCAUUCACCACAUCGAAAUCGAGCAGAGGCUUGCCGACCAGCGCAAUCUCAUCGAUUUUGUGGUGAAUCUUGGUUAUCUUCCCAUC ((((((((((((........))).)))))))))...((((((((((...(((((((...((((.(((((.....).)))))))).)))))))))))))))))..(((........))).. ( -45.00) >DroYak_CAF1 18562 115 - 1 ----AGGAAGAGUCCUCUGCCUCAU-CUUCCUCUUCAUCCACCACAUCAAAAUCGAGGAGAGGCUUGCCGACCAGCGCAAUCUCAUCGAUUUUAUGGUGAAUCUUGGUUAUCUUGCCAUC ----(((((((.............)-)))))).......(((((....((((((((.((((...(((((.....).)))))))).)))))))).))))).....((((......)))).. ( -32.22) >DroMoj_CAF1 29337 112 - 1 -------UGCUGCCAUCAUCUUCUU-CCUCCUCCGCAUCCGCCACCUCGAACUCGAGCAGUGGUUUGCCCACCAGAGCCAGCUCAUCGAUGUUGUGGUAGAUGCGCGUAAUCUUGCCAUC -------..(((((((((((.....-........(((...(((((((((....))))..))))).)))......(((....)))...))))..)))))))(((.(((......)))))). ( -33.20) >DroPer_CAF1 28287 115 - 1 -AGGAUUCAGUGCUAUU---CUCGU-CACCCUCCUCGUCGAGCACAUCAAAGUCCAGCAGGGGCUUGCCCACCAAUGCGAUCUCAUCGAUGCUGUGGUGGAUUUUAGUGAUUUUGCCAUC -.((.....(((((...---..((.-.........))...)))))..((((((((.((((....))))((((((..((.(((.....)))))..))))))......).)))))))))... ( -30.60) >consensus _AGGAAGAAGAGCCAUC_GCCUCAU_CUUCCUCCUCAUCCACCACAUCAAAAUCGAGCAGAGGCUUGCCCACCAGAGCAAUCUCAUCGAUGUUGUGGUGAAUCUUGGUUAUCUUGCCAUC ....................................(((.((((((.....(((((...((((.((((........)))))))).)))))..)))))).))).................. (-13.74 = -14.38 + 0.64)


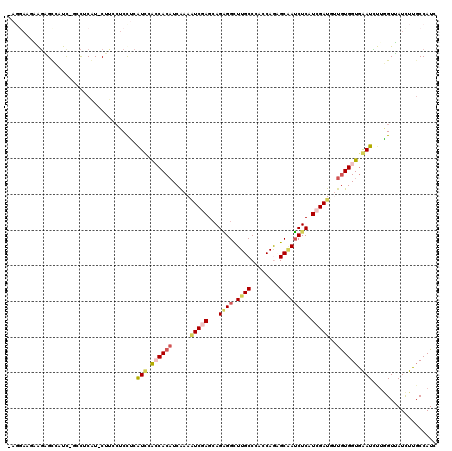
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:34 2006