
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,895,513 – 14,895,737 |
| Length | 224 |
| Max. P | 0.994101 |

| Location | 14,895,513 – 14,895,631 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 94.92 |
| Mean single sequence MFE | -34.54 |
| Consensus MFE | -29.62 |
| Energy contribution | -29.30 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14895513 118 + 22224390 GUCCUGGCGAAGGACUGGCAUUUGUUUUUGUUUGCCUACAGGAAGCUUUUAGCUCUUAUUGGCUCUUAUCAGCCUGCAAGCUUUUAUCCUGGAUGGCGGUCGUUAUUUUUGGACGACA (((((.....))))).((((........))))((((..((((((((((...((.......((((......)))).)))))))....)))))...))))((((((.......)))))). ( -35.50) >DroSec_CAF1 41484 118 + 1 GUCCUGGCGAAGGACUUGCAUUUGUUUUUGUUUGCCUACAGGAAGCUUUUCGCUCUUAUUGGCUCUUAUCAGCCUGCAAGCUUUUAUCCUGGAUGGCGGUCGUUAUUUUUGGACGACA (((((.....))))).................((((..((((((((((...((.......((((......)))).)))))))....)))))...))))((((((.......)))))). ( -35.10) >DroSim_CAF1 31977 118 + 1 GUCCUGGCGAAGGACUUGCAUUUGUUUUUGUUUGCCUACAGGAAGCUUUUCGCUCUUAUUGGCUCUUAUCAGCCUGCAAGCUUUUAUCCUGGAUGGCGGUCCUUAUUUUUGGACGACA ((((.....(((((((.(((........)))..(((..((((((((((...((.......((((......)))).)))))))....)))))...))))))))))......)))).... ( -36.80) >DroEre_CAF1 40328 117 + 1 GUCCUGGCGAAGGAU-UGCCUUUGUUUUUGUUUGCCUACAGGAAGCUUUUCGCUCUUAUUGGCUCUUAUCAGCUUGUAAGCUUUUAUCCCGGAUGACGGUCGUUAUUUUUGGACACCA ((((.((((((.((.-.((....))..)).))))))..(.((((((.....)))(((((.((((......)))).)))))......))).)((((((....))))))...)))).... ( -30.30) >DroYak_CAF1 40618 118 + 1 GUCCUGGCGAAGGAUUUGCAUUUGUUUUUGUUUGCCUAUAGGAAGCUUUUCGCUCUUAUUGGCUCUUAUCAGCCUGUAAGCUUUCAUCCUGGAUGGCGGUCGUUAUUUUUGGACGACA ((((.((.(((((.((((((...(((..((...(((.((((((.((.....)))))))).)))...))..))).)))))))))))..)).))))....((((((.......)))))). ( -35.00) >consensus GUCCUGGCGAAGGACUUGCAUUUGUUUUUGUUUGCCUACAGGAAGCUUUUCGCUCUUAUUGGCUCUUAUCAGCCUGCAAGCUUUUAUCCUGGAUGGCGGUCGUUAUUUUUGGACGACA ((((.((((((.((...((....))..)).))))))..((((((((((...((.......((((......)))).)))))))....)))))((((((....))))))...)))).... (-29.62 = -29.30 + -0.32)

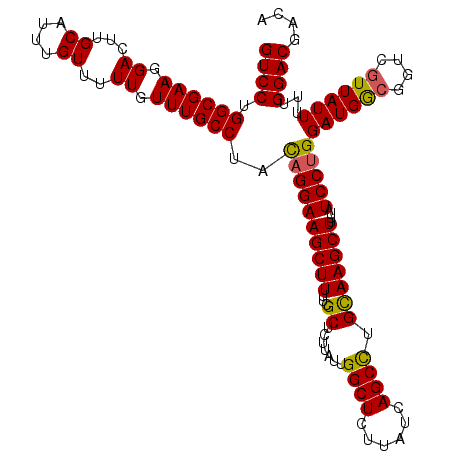

| Location | 14,895,513 – 14,895,631 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 94.92 |
| Mean single sequence MFE | -28.72 |
| Consensus MFE | -24.28 |
| Energy contribution | -24.76 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14895513 118 - 22224390 UGUCGUCCAAAAAUAACGACCGCCAUCCAGGAUAAAAGCUUGCAGGCUGAUAAGAGCCAAUAAGAGCUAAAAGCUUCCUGUAGGCAAACAAAAACAAAUGCCAGUCCUUCGCCAGGAC .(((((.........))))).(((...(((((....(((((...((((......)))).....))))).......)))))..)))..................(((((.....))))) ( -30.10) >DroSec_CAF1 41484 118 - 1 UGUCGUCCAAAAAUAACGACCGCCAUCCAGGAUAAAAGCUUGCAGGCUGAUAAGAGCCAAUAAGAGCGAAAAGCUUCCUGUAGGCAAACAAAAACAAAUGCAAGUCCUUCGCCAGGAC .(((((.........))))).(((...(((((......((((..((((......))))..))))(((.....))))))))..)))..................(((((.....))))) ( -30.20) >DroSim_CAF1 31977 118 - 1 UGUCGUCCAAAAAUAAGGACCGCCAUCCAGGAUAAAAGCUUGCAGGCUGAUAAGAGCCAAUAAGAGCGAAAAGCUUCCUGUAGGCAAACAAAAACAAAUGCAAGUCCUUCGCCAGGAC (((.((((........)))).(((...(((((......((((..((((......))))..))))(((.....))))))))..)))..............))).(((((.....))))) ( -31.40) >DroEre_CAF1 40328 117 - 1 UGGUGUCCAAAAAUAACGACCGUCAUCCGGGAUAAAAGCUUACAAGCUGAUAAGAGCCAAUAAGAGCGAAAAGCUUCCUGUAGGCAAACAAAAACAAAGGCA-AUCCUUCGCCAGGAC ....((((...........(((.....))).......(((((((.(((......)))......((((.....))))..))))))).............(((.-.......))).)))) ( -26.70) >DroYak_CAF1 40618 118 - 1 UGUCGUCCAAAAAUAACGACCGCCAUCCAGGAUGAAAGCUUACAGGCUGAUAAGAGCCAAUAAGAGCGAAAAGCUUCCUAUAGGCAAACAAAAACAAAUGCAAAUCCUUCGCCAGGAC .(((((.........))))).(((....((((......((((..((((......))))..))))(((.....)))))))...)))...................((((.....)))). ( -25.20) >consensus UGUCGUCCAAAAAUAACGACCGCCAUCCAGGAUAAAAGCUUGCAGGCUGAUAAGAGCCAAUAAGAGCGAAAAGCUUCCUGUAGGCAAACAAAAACAAAUGCAAGUCCUUCGCCAGGAC .(((((.........))))).(((...(((((......((((..((((......))))..))))(((.....))))))))..)))..................(((((.....))))) (-24.28 = -24.76 + 0.48)



| Location | 14,895,553 – 14,895,671 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 93.22 |
| Mean single sequence MFE | -31.60 |
| Consensus MFE | -27.44 |
| Energy contribution | -27.96 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.87 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14895553 118 - 22224390 GCACCUUGGUGUCCGAUAUGUGUACAUAUGCAUGUGCAUCUGUCGUCCAAAAAUAACGACCGCCAUCCAGGAUAAAAGCUUGCAGGCUGAUAAGAGCCAAUAAGAGCUAAAAGCUUCC ((.((((((((...((((.((((((((....)))))))).))))(((..........))))))))...))).....(((((...((((......)))).....)))))....)).... ( -33.20) >DroSec_CAF1 41524 118 - 1 GCACCUUGGUGCCCGAUAUGUGUACAUAUGCAUGUGCAUCUGUCGUCCAAAAAUAACGACCGCCAUCCAGGAUAAAAGCUUGCAGGCUGAUAAGAGCCAAUAAGAGCGAAAAGCUUCC ((.((((((((...((((.((((((((....)))))))).))))(((..........))))))))...)))......((((...((((......)))).....)))).....)).... ( -32.30) >DroSim_CAF1 32017 118 - 1 GCACCUUGGUGCCCGAUAUGUGUACAUAUGCAUGUGCAUCUGUCGUCCAAAAAUAAGGACCGCCAUCCAGGAUAAAAGCUUGCAGGCUGAUAAGAGCCAAUAAGAGCGAAAAGCUUCC ((.((((((((...((((.((((((((....)))))))).))))((((........)))))))))...)))......((((...((((......)))).....)))).....)).... ( -37.10) >DroEre_CAF1 40367 118 - 1 GCACCUUGGUGGGCGAUAUGUGUACAUAUGCAUGUGCAUCUGGUGUCCAAAAAUAACGACCGUCAUCCGGGAUAAAAGCUUACAAGCUGAUAAGAGCCAAUAAGAGCGAAAAGCUUCC .....((((((((((.((.((((((((....)))))))).)).)))))...........(((.....)))......((((....)))).......)))))...((((.....)))).. ( -28.60) >DroYak_CAF1 40658 112 - 1 GCACCUUGGUGCUCGAUAUGUGUUC------AUGUGCAUCUGUCGUCCAAAAAUAACGACCGCCAUCCAGGAUGAAAGCUUACAGGCUGAUAAGAGCCAAUAAGAGCGAAAAGCUUCC ((((....))))...(((((....)------))))((....(((((.........)))))(((((((...))))....((((..((((......))))..)))).)))....)).... ( -26.80) >consensus GCACCUUGGUGCCCGAUAUGUGUACAUAUGCAUGUGCAUCUGUCGUCCAAAAAUAACGACCGCCAUCCAGGAUAAAAGCUUGCAGGCUGAUAAGAGCCAAUAAGAGCGAAAAGCUUCC ((.((((((((...((((.((((((((....)))))))).))))(((..........))))))))...)))......((((...((((......)))).....)))).....)).... (-27.44 = -27.96 + 0.52)

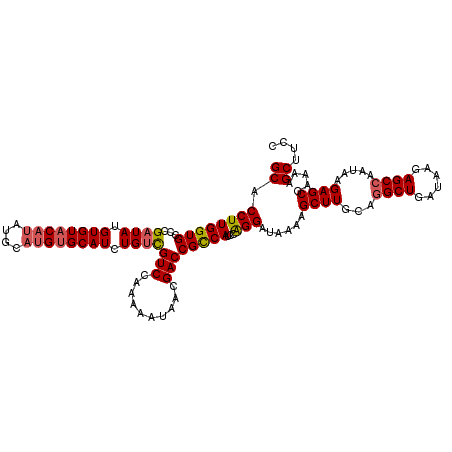
| Location | 14,895,591 – 14,895,698 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.62 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -21.62 |
| Energy contribution | -21.94 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.869786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14895591 107 - 22224390 -----UGCAUA--------UAUAUGUGCAUCCCAAAGCUUGCACCUUGGUGUCCGAUAUGUGUACAUAUGCAUGUGCAUCUGUCGUCCAAAAAUAACGACCGCCAUCCAGGAUAAAAGCU -----((((((--------....))))))......(((((...((((((((...((((.((((((((....)))))))).))))(((..........))))))))...)))....))))) ( -29.90) >DroSec_CAF1 41562 120 - 1 GAGUAUGCAUACAUAUAUACAUAUGUGCAUCCCAAAGCUUGCACCUUGGUGCCCGAUAUGUGUACAUAUGCAUGUGCAUCUGUCGUCCAAAAAUAACGACCGCCAUCCAGGAUAAAAGCU .((.((((((((((((.(((((((((((........))..((((....))))...))))))))).)))))..)))))))))(((((.........)))))..((.....))......... ( -31.10) >DroSim_CAF1 32055 120 - 1 GAGUAUGCAUAUAUAUAUACAUAUGUGCAUCCCAAAGCUUGCACCUUGGUGCCCGAUAUGUGUACAUAUGCAUGUGCAUCUGUCGUCCAAAAAUAAGGACCGCCAUCCAGGAUAAAAGCU (.(.((((((((((......)))))))))).))..(((((...((((((((...((((.((((((((....)))))))).))))((((........)))))))))...)))....))))) ( -39.30) >DroEre_CAF1 40405 112 - 1 -----UACA---ACAUACAUGUAUGUACAUCCCAAAGCUUGCACCUUGGUGGGCGAUAUGUGUACAUAUGCAUGUGCAUCUGGUGUCCAAAAAUAACGACCGUCAUCCGGGAUAAAAGCU -----....---(((((....)))))..(((((...(((..(......)..)))(((((((((((((....))))))))...))))).....................)))))....... ( -28.60) >DroYak_CAF1 40696 102 - 1 -----UACA----UAUAC---UAUGUAGACCCAAAAGCUUGCACCUUGGUGCUCGAUAUGUGUUC------AUGUGCAUCUGUCGUCCAAAAAUAACGACCGCCAUCCAGGAUGAAAGCU -----((((----((...---))))))........(((((.((((((((((.((((((.((((..------....)))).))))((.........)))).)))))...))).)).))))) ( -23.50) >consensus _____UGCAUA_AUAUACACAUAUGUGCAUCCCAAAGCUUGCACCUUGGUGCCCGAUAUGUGUACAUAUGCAUGUGCAUCUGUCGUCCAAAAAUAACGACCGCCAUCCAGGAUAAAAGCU ...................................(((((...((((((((...((((.((((((((....)))))))).))))(((..........))))))))...)))....))))) (-21.62 = -21.94 + 0.32)

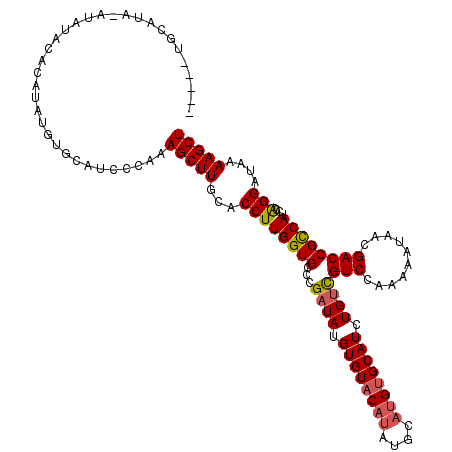
| Location | 14,895,631 – 14,895,737 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.62 |
| Mean single sequence MFE | -35.09 |
| Consensus MFE | -16.13 |
| Energy contribution | -16.41 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.46 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14895631 106 + 22224390 GAUGCACAUGCAUAUGUACACAUAUCGGACACCAAGGUGCAAGCUUUGGGAUGCACAUAUA--------UAUGCA------UAUACCCACAUGACCAUCUUUGCUACUGAGUGCAUCCGG ((((((((((((((((((....((((.....(((((((....)))))))))))....))))--------))))))------)...............((.........)))))))))... ( -29.90) >DroSec_CAF1 41602 120 + 1 GAUGCACAUGCAUAUGUACACAUAUCGGGCACCAAGGUGCAAGCUUUGGGAUGCACAUAUGUAUAUAUGUAUGCAUACUCGUAUACCCACAUGACCAUUUUUGCGACUGAAUGCAUCCGG ((((((((((((((((((((..(((...((((((((((....)))))))..)))..))))))))))))))))).....(((((..................))))).....))))))... ( -36.77) >DroSim_CAF1 32095 120 + 1 GAUGCACAUGCAUAUGUACACAUAUCGGGCACCAAGGUGCAAGCUUUGGGAUGCACAUAUGUAUAUAUAUAUGCAUACUCGUACACCCACAUGACCAUUUUUGCGACUGAAUGCAUCCGG ((((((.(((((((((((.((((((...((((((((((....)))))))..)))..))))))...)))))))))))..(((((..................))))).....))))))... ( -36.17) >DroEre_CAF1 40445 111 + 1 GAUGCACAUGCAUAUGUACACAUAUCGCCCACCAAGGUGCAAGCUUUGGGAUGUACAUACAUGUAUGU---UGUA------CCCAGACAUGUGACCAUUUAUGUGACUGUGUGCAUCCGA (((((((((.((((((..(((((.((((((.....)).))......((((....(((((....)))))---....------)))))).)))))......))))))...)))))))))... ( -36.70) >DroYak_CAF1 40736 101 + 1 GAUGCACAU------GAACACAUAUCGAGCACCAAGGUGCAAGCUUUUGGGUCUACAUA---GUAUA----UGUA------CAUACACAUGUGACCACUGAUGCGAGCGUGUGCAUCCGG (((((((((------(..(.........((((....))))..((.((..((((.((((.---((.((----(...------.))).))))))))))...)).)))..))))))))))... ( -35.90) >consensus GAUGCACAUGCAUAUGUACACAUAUCGGGCACCAAGGUGCAAGCUUUGGGAUGCACAUAUAUAUAUAU_UAUGCA______UAUACCCACAUGACCAUUUUUGCGACUGAGUGCAUCCGG (((((((.....................((((....))))..((...((((((..(((................................)))..)))))).))......)))))))... (-16.13 = -16.41 + 0.28)


| Location | 14,895,631 – 14,895,737 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.62 |
| Mean single sequence MFE | -35.40 |
| Consensus MFE | -20.80 |
| Energy contribution | -20.34 |
| Covariance contribution | -0.46 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14895631 106 - 22224390 CCGGAUGCACUCAGUAGCAAAGAUGGUCAUGUGGGUAUA------UGCAUA--------UAUAUGUGCAUCCCAAAGCUUGCACCUUGGUGUCCGAUAUGUGUACAUAUGCAUGUGCAUC .(((..(((((.((..(((.....(((....((((...(------((((((--------....)))))))))))..))))))..)).))))))))....((((((((....)))))))). ( -31.90) >DroSec_CAF1 41602 120 - 1 CCGGAUGCAUUCAGUCGCAAAAAUGGUCAUGUGGGUAUACGAGUAUGCAUACAUAUAUACAUAUGUGCAUCCCAAAGCUUGCACCUUGGUGCCCGAUAUGUGUACAUAUGCAUGUGCAUC ...(((((....................(((((.(((((....))))).)))))((((.(((((((((((.......(..((((....))))..)....))))))))))).))))))))) ( -33.30) >DroSim_CAF1 32095 120 - 1 CCGGAUGCAUUCAGUCGCAAAAAUGGUCAUGUGGGUGUACGAGUAUGCAUAUAUAUAUACAUAUGUGCAUCCCAAAGCUUGCACCUUGGUGCCCGAUAUGUGUACAUAUGCAUGUGCAUC .((.((((((...((((((....(((.(((..((((((..((((((((((((((......))))))))))......))))))))))..))).)))...)))).))..)))))).)).... ( -36.60) >DroEre_CAF1 40445 111 - 1 UCGGAUGCACACAGUCACAUAAAUGGUCACAUGUCUGGG------UACA---ACAUACAUGUAUGUACAUCCCAAAGCUUGCACCUUGGUGGGCGAUAUGUGUACAUAUGCAUGUGCAUC ...((((((((......((((..((..((((((((((((------....---(((((....)))))....))))..(((..(......)..)))))))))))..))))))..)))))))) ( -38.80) >DroYak_CAF1 40736 101 - 1 CCGGAUGCACACGCUCGCAUCAGUGGUCACAUGUGUAUG------UACA----UAUAC---UAUGUAGACCCAAAAGCUUGCACCUUGGUGCUCGAUAUGUGUUC------AUGUGCAUC ...((((((((.(..(((((..(.((((((((((((((.------...)----)))))---.)))).)))))........((((....)))).....)))))..)------.)))))))) ( -36.40) >consensus CCGGAUGCACUCAGUCGCAAAAAUGGUCAUGUGGGUAUA______UGCAUA_AUAUACACAUAUGUGCAUCCCAAAGCUUGCACCUUGGUGCCCGAUAUGUGUACAUAUGCAUGUGCAUC ...((((((((((((.((.......)).)).))).........................(((((((((((.......(..((((....))))..)....)))))))))))...))))))) (-20.80 = -20.34 + -0.46)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:31 2006