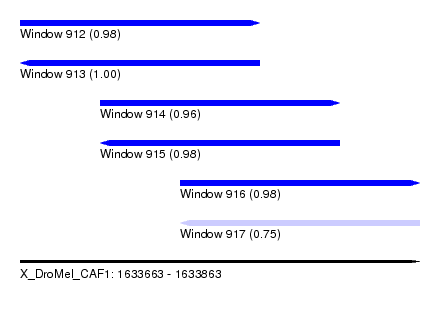
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,633,663 – 1,633,863 |
| Length | 200 |
| Max. P | 0.995429 |

| Location | 1,633,663 – 1,633,783 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.61 |
| Mean single sequence MFE | -29.25 |
| Consensus MFE | -26.56 |
| Energy contribution | -27.12 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1633663 120 + 22224390 GCAUGGCACAUGUGCCUUCCCUUUAAAAGGGUAUAUAAAUAUCGAGCUCCUCGAGAUCGCAGCUAGUCGCGCAAAUUAUCUGCACACUUACAAACAUGUGUUUUUACACAUGUAUGUACA ....((((....))))..((((.....))))..........(((((...)))))....((((.((((.......)))).)))).....((((.((((((((....)))))))).)))).. ( -33.60) >DroSim_CAF1 45372 115 + 1 G-----CACAUGUGCCUUCCCUUUAAAAGGGUAUAUAAAUAUCGAGCUCCUCGAGAUCGCAGCUAGUCGCGCAAAUUAUCUGCACACUUACAAACAUGUGUUUUUACACAUGUAUGUACA .-----...(((((((((.........))))))))).....(((((...)))))....((((.((((.......)))).)))).....((((.((((((((....)))))))).)))).. ( -30.90) >DroEre_CAF1 29944 115 + 1 C-----CACAUGCGCCUUCCCUUUAAAAGGGUAUAUAAAUAUCGAGCUCUUCGAAAUCGCAGCUAGUCGCGCAAAUUAUCUGCACACUUACAAACUUGUGUUUUUACACAUGUAUGUACA .-----....(((((...((((.....)))).............((((...((....)).))))....)))))...............((((.((.(((((....))))).)).)))).. ( -25.20) >DroYak_CAF1 33245 115 + 1 C-----CACAUGUGCCUUCCCUUUAAAAGGGUAUAUAAAUAUCGAGCUCCUCAAAAUCGCUGCUAGUCGCGCAAAUUAUCUGCACACUUACAAACAUGUGUUUUUACACAUGUAUGUACA .-----....(((((...((((.....))))............(((...)))......((.((.....)))).........)))))..((((.((((((((....)))))))).)))).. ( -27.30) >consensus C_____CACAUGUGCCUUCCCUUUAAAAGGGUAUAUAAAUAUCGAGCUCCUCGAAAUCGCAGCUAGUCGCGCAAAUUAUCUGCACACUUACAAACAUGUGUUUUUACACAUGUAUGUACA ..........(((((...((((.....))))..........(((((...)))))....((.((.....)))).........)))))..((((.((((((((....)))))))).)))).. (-26.56 = -27.12 + 0.56)

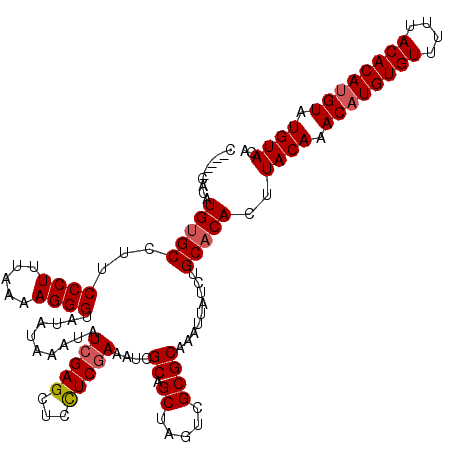
| Location | 1,633,663 – 1,633,783 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.61 |
| Mean single sequence MFE | -35.22 |
| Consensus MFE | -34.75 |
| Energy contribution | -34.88 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.995429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1633663 120 - 22224390 UGUACAUACAUGUGUAAAAACACAUGUUUGUAAGUGUGCAGAUAAUUUGCGCGACUAGCUGCGAUCUCGAGGAGCUCGAUAUUUAUAUACCCUUUUAAAGGGAAGGCACAUGUGCCAUGC ..((((.((((((((....)))))))).))))((((((((((...))))))).)))((((.(........).)))).............((((.....))))..((((....)))).... ( -38.30) >DroSim_CAF1 45372 115 - 1 UGUACAUACAUGUGUAAAAACACAUGUUUGUAAGUGUGCAGAUAAUUUGCGCGACUAGCUGCGAUCUCGAGGAGCUCGAUAUUUAUAUACCCUUUUAAAGGGAAGGCACAUGUG-----C ..((((.((((((((....)))))))).)))).(((((((((((..((((((.....)).))))..(((((...)))))))))).....((((.....))))...))))))...-----. ( -36.90) >DroEre_CAF1 29944 115 - 1 UGUACAUACAUGUGUAAAAACACAAGUUUGUAAGUGUGCAGAUAAUUUGCGCGACUAGCUGCGAUUUCGAAGAGCUCGAUAUUUAUAUACCCUUUUAAAGGGAAGGCGCAUGUG-----G ..((((.((.(((((....))))).)).)))).(((((((((((..((((((.....)).))))..((((.....))))))))).....((((.....))))...))))))...-----. ( -30.80) >DroYak_CAF1 33245 115 - 1 UGUACAUACAUGUGUAAAAACACAUGUUUGUAAGUGUGCAGAUAAUUUGCGCGACUAGCAGCGAUUUUGAGGAGCUCGAUAUUUAUAUACCCUUUUAAAGGGAAGGCACAUGUG-----G ..((((.((((((((....)))))))).)))).(((((((((((..((((((.....)).))))..(((((...)))))))))).....((((.....))))...))))))...-----. ( -34.90) >consensus UGUACAUACAUGUGUAAAAACACAUGUUUGUAAGUGUGCAGAUAAUUUGCGCGACUAGCUGCGAUCUCGAGGAGCUCGAUAUUUAUAUACCCUUUUAAAGGGAAGGCACAUGUG_____C ..((((.((((((((....)))))))).)))).(((((((((((..((((((.....)).))))..(((((...)))))))))).....((((.....))))...))))))......... (-34.75 = -34.88 + 0.12)

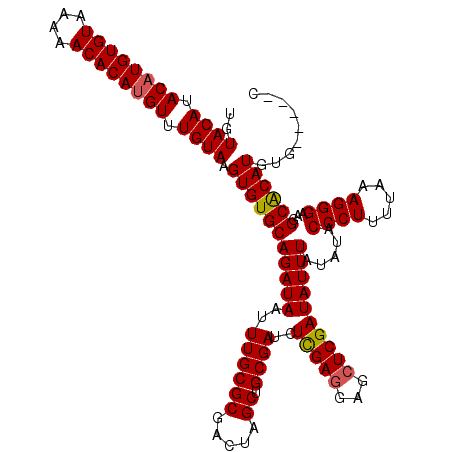
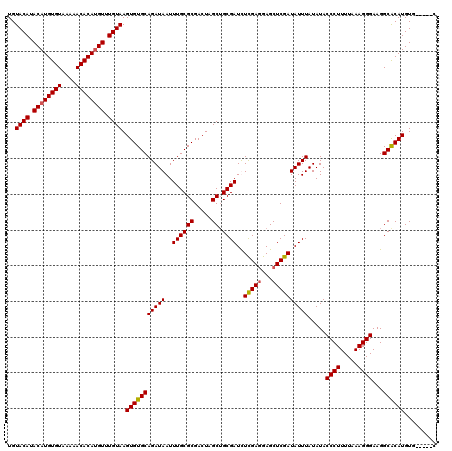
| Location | 1,633,703 – 1,633,823 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.39 |
| Mean single sequence MFE | -23.02 |
| Consensus MFE | -21.31 |
| Energy contribution | -21.62 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1633703 120 + 22224390 AUCGAGCUCCUCGAGAUCGCAGCUAGUCGCGCAAAUUAUCUGCACACUUACAAACAUGUGUUUUUACACAUGUAUGUACAUUUUUAAAAUUUUUUUUUCAUAAUUGCAGUUCCUUCUACA .(((((...)))))....((.((.....)))).......(((((....((((.((((((((....)))))))).)))).......((((....)))).......)))))........... ( -24.70) >DroSim_CAF1 45407 118 + 1 AUCGAGCUCCUCGAGAUCGCAGCUAGUCGCGCAAAUUAUCUGCACACUUACAAACAUGUGUUUUUACACAUGUAUGUACAUUUUUUAAAUUU--UUUUCAUAAUUGCAGUUCCUUCUACA .(((((...)))))....((.((.....)))).......((((.....((((.((((((((....)))))))).)))).........((((.--.......))))))))........... ( -24.70) >DroEre_CAF1 29979 120 + 1 AUCGAGCUCUUCGAAAUCGCAGCUAGUCGCGCAAAUUAUCUGCACACUUACAAACUUGUGUUUUUACACAUGUAUGUACAUUUUUAAAAUUUUUUUUUCAUAAUUGCAGUUCCUUCUACA ...((((((...((((..((((.((((.......)))).)))).....((((.((.(((((....))))).)).)))).................))))......).)))))........ ( -19.10) >DroYak_CAF1 33280 120 + 1 AUCGAGCUCCUCAAAAUCGCUGCUAGUCGCGCAAAUUAUCUGCACACUUACAAACAUGUGUUUUUACACAUGUAUGUACAUUUUUUAAAUUUUUUUUUCAUAAUUGCAGUUCCUUCUACA ...(((...)))......(((((.(((.(.(((.......))).))))((((.((((((((....)))))))).))))...........................))))).......... ( -23.60) >consensus AUCGAGCUCCUCGAAAUCGCAGCUAGUCGCGCAAAUUAUCUGCACACUUACAAACAUGUGUUUUUACACAUGUAUGUACAUUUUUAAAAUUUUUUUUUCAUAAUUGCAGUUCCUUCUACA ...(((((...(((....((((.((((.......)))).)))).....((((.((((((((....)))))))).)))).........................))).)))))........ (-21.31 = -21.62 + 0.31)

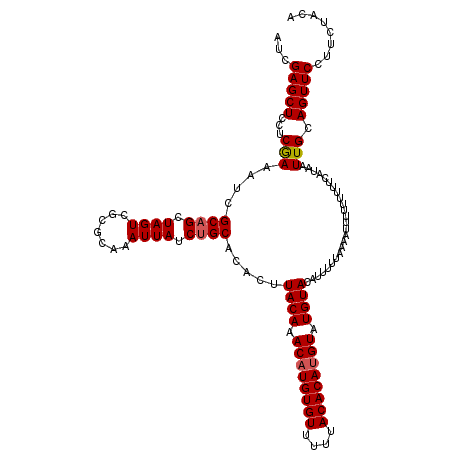
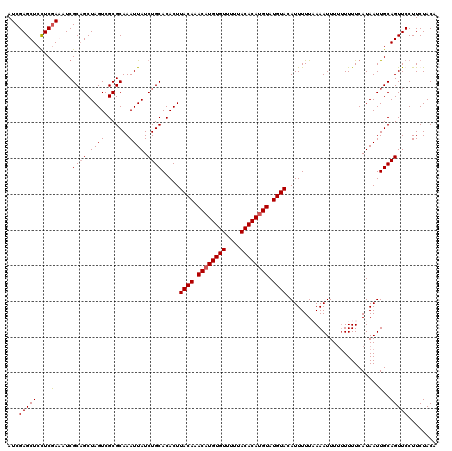
| Location | 1,633,703 – 1,633,823 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.39 |
| Mean single sequence MFE | -28.85 |
| Consensus MFE | -27.14 |
| Energy contribution | -26.95 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1633703 120 - 22224390 UGUAGAAGGAACUGCAAUUAUGAAAAAAAAAUUUUAAAAAUGUACAUACAUGUGUAAAAACACAUGUUUGUAAGUGUGCAGAUAAUUUGCGCGACUAGCUGCGAUCUCGAGGAGCUCGAU .((((..((.((((((.((.(((((......))))).)).)))(((.((((((((....)))))))).))).)))(((((((...)))))))..))..))))....(((((...))))). ( -30.60) >DroSim_CAF1 45407 118 - 1 UGUAGAAGGAACUGCAAUUAUGAAAA--AAAUUUAAAAAAUGUACAUACAUGUGUAAAAACACAUGUUUGUAAGUGUGCAGAUAAUUUGCGCGACUAGCUGCGAUCUCGAGGAGCUCGAU (((((......)))))..........--..............((((.((((((((....)))))))).))))((((((((((...))))))).)))((((.(........).)))).... ( -30.40) >DroEre_CAF1 29979 120 - 1 UGUAGAAGGAACUGCAAUUAUGAAAAAAAAAUUUUAAAAAUGUACAUACAUGUGUAAAAACACAAGUUUGUAAGUGUGCAGAUAAUUUGCGCGACUAGCUGCGAUUUCGAAGAGCUCGAU (((((......)))))..........................((((.((.(((((....))))).)).))))((((((((((...))))))).)))((((.((....))...)))).... ( -25.10) >DroYak_CAF1 33280 120 - 1 UGUAGAAGGAACUGCAAUUAUGAAAAAAAAAUUUAAAAAAUGUACAUACAUGUGUAAAAACACAUGUUUGUAAGUGUGCAGAUAAUUUGCGCGACUAGCAGCGAUUUUGAGGAGCUCGAU ...........((((...........................((((.((((((((....)))))))).))))((((((((((...))))))).))).)))).....(((((...))))). ( -29.30) >consensus UGUAGAAGGAACUGCAAUUAUGAAAAAAAAAUUUAAAAAAUGUACAUACAUGUGUAAAAACACAUGUUUGUAAGUGUGCAGAUAAUUUGCGCGACUAGCUGCGAUCUCGAGGAGCUCGAU (((((......)))))..........................((((.((((((((....)))))))).))))((((((((((...))))))).))).....(((.(((...))).))).. (-27.14 = -26.95 + -0.19)

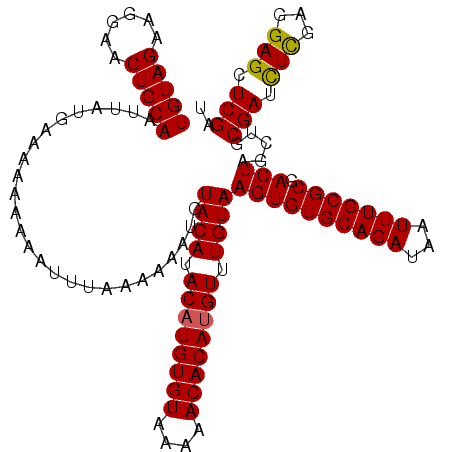
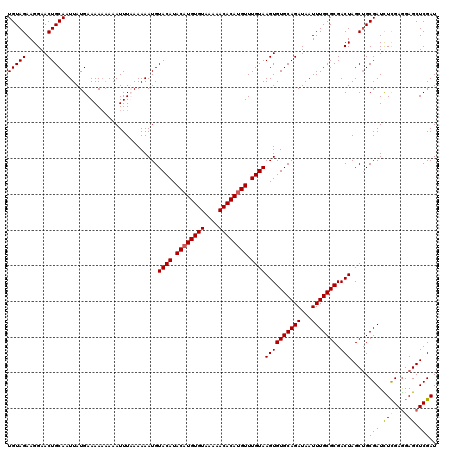
| Location | 1,633,743 – 1,633,863 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.19 |
| Mean single sequence MFE | -27.93 |
| Consensus MFE | -27.95 |
| Energy contribution | -28.20 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.28 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1633743 120 + 22224390 UGCACACUUACAAACAUGUGUUUUUACACAUGUAUGUACAUUUUUAAAAUUUUUUUUUCAUAAUUGCAGUUCCUUCUACAAAAUUAGCUGCAGUUGGCUUUUGUCGUUGUCAGAUAAAUG .((.....((((.((((((((....)))))))).))))......................(((((((((((..((......))..))))))))))))).((((((.......)))))).. ( -29.00) >DroSim_CAF1 45447 118 + 1 UGCACACUUACAAACAUGUGUUUUUACACAUGUAUGUACAUUUUUUAAAUUU--UUUUCAUAAUUGCAGUUCCUUCUACAAAAUUAGCUGCAGUUGGCUUUUGUCGUUGUCAGAUAAAUG .((.....((((.((((((((....)))))))).))))..............--......(((((((((((..((......))..))))))))))))).((((((.......)))))).. ( -29.00) >DroEre_CAF1 30019 120 + 1 UGCACACUUACAAACUUGUGUUUUUACACAUGUAUGUACAUUUUUAAAAUUUUUUUUUCAUAAUUGCAGUUCCUUCUACAAAAUUAGCUGCAGUUGGCUUUUGUCGUUGUCAGAUAAAUG .((.....((((.((.(((((....))))).)).))))......................(((((((((((..((......))..))))))))))))).((((((.......)))))).. ( -24.70) >DroYak_CAF1 33320 120 + 1 UGCACACUUACAAACAUGUGUUUUUACACAUGUAUGUACAUUUUUUAAAUUUUUUUUUCAUAAUUGCAGUUCCUUCUACAAAAUUAGCUGCAGUUGGCUUUUGUCGUUGUCAGAUAAAUG .((.....((((.((((((((....)))))))).))))......................(((((((((((..((......))..))))))))))))).((((((.......)))))).. ( -29.00) >consensus UGCACACUUACAAACAUGUGUUUUUACACAUGUAUGUACAUUUUUAAAAUUUUUUUUUCAUAAUUGCAGUUCCUUCUACAAAAUUAGCUGCAGUUGGCUUUUGUCGUUGUCAGAUAAAUG .((.....((((.((((((((....)))))))).))))......................(((((((((((..((......))..))))))))))))).((((((.......)))))).. (-27.95 = -28.20 + 0.25)

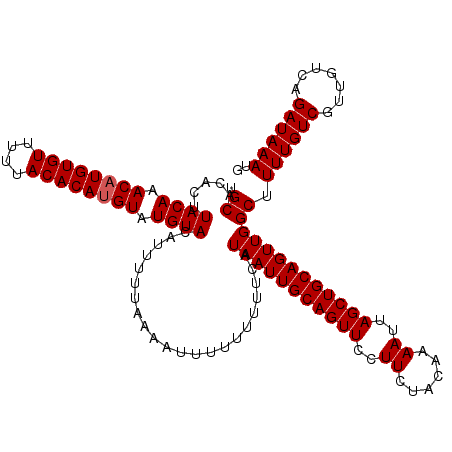
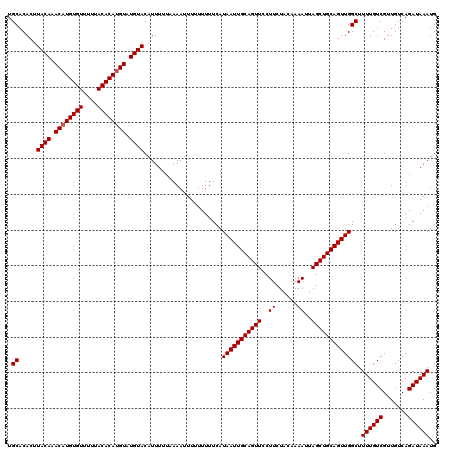
| Location | 1,633,743 – 1,633,863 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.19 |
| Mean single sequence MFE | -22.00 |
| Consensus MFE | -21.55 |
| Energy contribution | -21.80 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.750152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1633743 120 - 22224390 CAUUUAUCUGACAACGACAAAAGCCAACUGCAGCUAAUUUUGUAGAAGGAACUGCAAUUAUGAAAAAAAAAUUUUAAAAAUGUACAUACAUGUGUAAAAACACAUGUUUGUAAGUGUGCA .......................((..((((((......))))))..))....(((....(((((......)))))......((((.((((((((....)))))))).))))....))). ( -23.40) >DroSim_CAF1 45447 118 - 1 CAUUUAUCUGACAACGACAAAAGCCAACUGCAGCUAAUUUUGUAGAAGGAACUGCAAUUAUGAAAA--AAAUUUAAAAAAUGUACAUACAUGUGUAAAAACACAUGUUUGUAAGUGUGCA .............................((((((....((((((......)))))).........--..............((((.((((((((....)))))))).))))))).))). ( -22.60) >DroEre_CAF1 30019 120 - 1 CAUUUAUCUGACAACGACAAAAGCCAACUGCAGCUAAUUUUGUAGAAGGAACUGCAAUUAUGAAAAAAAAAUUUUAAAAAUGUACAUACAUGUGUAAAAACACAAGUUUGUAAGUGUGCA (((((((..(((...........((..((((((......))))))..))...((((.((.(((((......))))).)).))))......(((((....))))).))).))))))).... ( -19.40) >DroYak_CAF1 33320 120 - 1 CAUUUAUCUGACAACGACAAAAGCCAACUGCAGCUAAUUUUGUAGAAGGAACUGCAAUUAUGAAAAAAAAAUUUAAAAAAUGUACAUACAUGUGUAAAAACACAUGUUUGUAAGUGUGCA .............................((((((....((((((......)))))).........................((((.((((((((....)))))))).))))))).))). ( -22.60) >consensus CAUUUAUCUGACAACGACAAAAGCCAACUGCAGCUAAUUUUGUAGAAGGAACUGCAAUUAUGAAAAAAAAAUUUAAAAAAUGUACAUACAUGUGUAAAAACACAUGUUUGUAAGUGUGCA .............................((((((....((((((......)))))).........................((((.((((((((....)))))))).))))))).))). (-21.55 = -21.80 + 0.25)

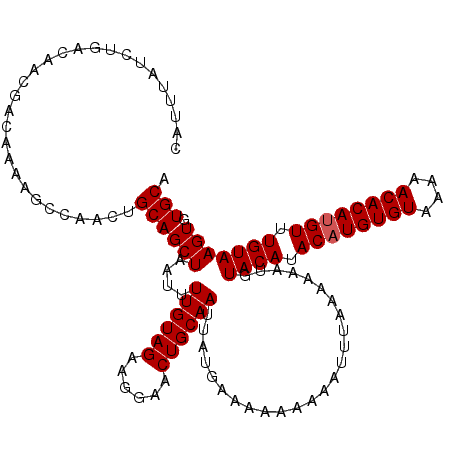
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:19 2006