
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,894,853 – 14,894,984 |
| Length | 131 |
| Max. P | 0.955819 |

| Location | 14,894,853 – 14,894,944 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 76.10 |
| Mean single sequence MFE | -37.78 |
| Consensus MFE | -19.76 |
| Energy contribution | -19.65 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
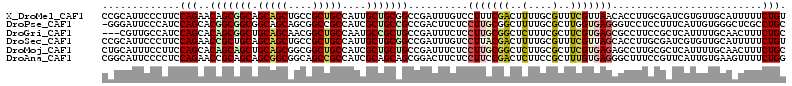
>X_DroMel_CAF1 14894853 91 - 22224390 CAAAAGUCGAAAGGACAAAUCGGCCGCAGCAAUGGCAGCGGCAGCUGCUGCCGCUGUUCUGGAAGGGAAUGCGG------GUGCCGG---GGGACUGGUG .....(((.....))).......(((((......((((((((((...))))))))))(((.....))).)))))------..(((((---....))))). ( -38.10) >DroEre_CAF1 39724 91 - 1 CAAAAGUCGGAAGGAUAAAUCGGCAGCAGCAAUGGCAGCGGCAGCUGCUGCAGCGGUUCUGGAGGGGAAUGCGG------GGGCCGG---GGGACUGGUG ......((....))........((((((((....((....)).)))))))).(((.((((....)))).)))..------..(((((---....))))). ( -33.20) >DroYak_CAF1 40004 91 - 1 CAAAAGUCGGAAGGACAAGUCGGCGGCCGCAAUGGCCGCGGCAGCUGCUGCAGCGGUUCUGGAGGGGAAUGCGG------GAGCUGG---UGGACUGGUG ......((....))((.((((.((((((.....))))))..(((((.(((((....((((....)))).)))))------.))))).---..)))).)). ( -38.40) >DroMoj_CAF1 51400 100 - 1 CAAGAGCCGCAAGGAGAAAUCGGCAGCAGCGAUGGCAGCCGCCGCUGCAGCUGCUGUGCUGGAAGGAAAUGCAGCUGGCGGUGCGGGAGCAGGCAUUGUA .....((.((...((....)).)).)).((((((.(.((((((((((((..(.((........)).)..)))))).))))))((....)).).)))))). ( -42.70) >DroAna_CAF1 54633 91 - 1 GAAGAGUCGGAAGGAGAAGUCCGCUGCUGCGAUGGCGGCUGCCGCCGCUGCUGCGGUUCUGGAGGGGAAUGCCG------GAGCCGG---AGGUCUAGUG ......((....))(((..(((((.((.(((.((((....)))).))).)).))((((((((.........)))------)))))))---)..))).... ( -37.80) >DroPer_CAF1 68339 85 - 1 CAAAAGUCGCAAGGAGAAGUCGGCGGCAGCGAUGGCGGCUGCUGCUGCCGCCGCCGUGCUGGAUGGGAAUCCC------------GG---UAGCAUUGUG .......(((((......(.((((((((((...(((....))))))))))))).).(((((..((((...)))------------).---)))))))))) ( -36.50) >consensus CAAAAGUCGGAAGGAGAAAUCGGCAGCAGCAAUGGCAGCGGCAGCUGCUGCAGCGGUUCUGGAGGGGAAUGCGG______GAGCCGG___AGGACUGGUG ......((....)).....((.(..(((((..(((((((....)))))))..)))))..).))..................................... (-19.76 = -19.65 + -0.11)



| Location | 14,894,870 – 14,894,984 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 76.01 |
| Mean single sequence MFE | -39.38 |
| Consensus MFE | -19.94 |
| Energy contribution | -20.42 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14894870 114 + 22224390 CCGCAUUCCCUUCCAGAACAGCGGCAGCAGCUGCCGCUGCCAUUGCUGCGGCCGAUUUGUCCUUUCGACUUUUGCGUUUCGUUAACACCUUGCGAUCGUGUUGCAUUUUUCUGU .............((((((((((((((...))))))))).......(((((((((((.(((.....)))....(((..............)))))))).))))))...))))). ( -34.04) >DroPse_CAF1 50271 113 + 1 -GGGAUUCCCAUCCAGCACGGCGGCGGCAGCAGCGGCCGCCAUCGCUGCCGCCGACUUCUCCUUGCGGCUUUUGCGCUUGGUGAGGGUCCUCCUUUCAUUGUGGGCUCGCCUGC -(((((((.((((.(((..((((((.((....)).))))))...((.(((((.((....))...)))))....))))).)))).))))))).........(..((....))..) ( -50.60) >DroGri_CAF1 49377 111 + 1 ---CGUUGCCAUCCAGCACAGCGGCUGCAGCAACGGCUGCCAAUGCCGCUGCCGAUUUCUCCUUGCGGCUCUUUCGCUUCGUGAGCGCCUUCCGCUCAUUUUGCAACUUUCUGC ---.(((((.....(((..((((((.(((((....)))))....))))))((((...........))))......)))..(((((((.....)))))))...)))))....... ( -42.20) >DroSec_CAF1 40866 114 + 1 CCGCAUUCCCUUCCAGAACCGCUGCAGCAGCUGCCGCUGCCAUUGCUGCGGCCGAUUUGUCCUUACGACUUUUGCGUUUCGUUAGCACCUUGCGAUCGUGUUGCAUUUUUCUGU .............((((((.((((((((((..((....))..)))))))))).)....(((.....)))...((((...((...((.....))...))...))))...))))). ( -31.00) >DroMoj_CAF1 51426 114 + 1 CUGCAUUUCCUUCCAGCACAGCAGCUGCAGCGGCGGCUGCCAUCGCUGCUGCCGAUUUCUCCUUGCGGCUCUUGCGCUUCGUGAGAGCCUUGCGCUCAUUUUGCAACUUUCUGC .((((.........(((.(((((((.(((((....)))))....))))))).............((((((((..((...))..))))))..))))).....))))......... ( -41.34) >DroAna_CAF1 54650 114 + 1 CGGCAUUCCCCUCCAGAACCGCAGCAGCGGCGGCAGCCGCCAUCGCAGCAGCGGACUUCUCCUUCCGACUCUUCCGCUUUGUGAGGGCUUUCCGUUCAUUGUGAAGUUUUCUGG .((......)).((((((((......(((((....)))))..((((((.((((((....((.....))....))))))))))))))((((..((.....))..)))).)))))) ( -37.10) >consensus CCGCAUUCCCUUCCAGAACAGCGGCAGCAGCAGCGGCUGCCAUCGCUGCGGCCGAUUUCUCCUUGCGACUCUUGCGCUUCGUGAGCGCCUUCCGAUCAUUUUGCAAUUUUCUGC .............(((..(((((((.(((((....)))))....)))))))..........(((((((..(....)..))))))).........................))). (-19.94 = -20.42 + 0.47)


| Location | 14,894,870 – 14,894,984 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 76.01 |
| Mean single sequence MFE | -43.03 |
| Consensus MFE | -24.00 |
| Energy contribution | -23.50 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
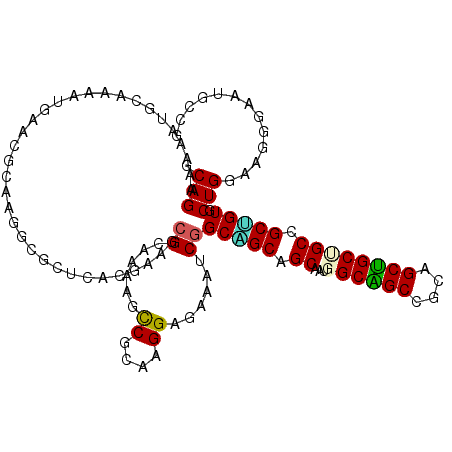
>X_DroMel_CAF1 14894870 114 - 22224390 ACAGAAAAAUGCAACACGAUCGCAAGGUGUUAACGAAACGCAAAAGUCGAAAGGACAAAUCGGCCGCAGCAAUGGCAGCGGCAGCUGCUGCCGCUGUUCUGGAAGGGAAUGCGG ............(((((...(....)))))).......((((...(((.....)))...((..(((.((((.(((((((((...))))))))).)))).)))..))...)))). ( -35.10) >DroPse_CAF1 50271 113 - 1 GCAGGCGAGCCCACAAUGAAAGGAGGACCCUCACCAAGCGCAAAAGCCGCAAGGAGAAGUCGGCGGCAGCGAUGGCGGCCGCUGCUGCCGCCGCCGUGCUGGAUGGGAAUCCC- ...((.((.((((.........(((....))).(((.((((.....((....))....(.(((((((((((.(((...))).))))))))))).)))))))).))))..))))- ( -51.00) >DroGri_CAF1 49377 111 - 1 GCAGAAAGUUGCAAAAUGAGCGGAAGGCGCUCACGAAGCGAAAGAGCCGCAAGGAGAAAUCGGCAGCGGCAUUGGCAGCCGUUGCUGCAGCCGCUGUGCUGGAUGGCAACG--- .......(((((....((((((.....))))))...(((((.....((....)).....)).((((((((....(((((....))))).))))))))))).....))))).--- ( -48.20) >DroSec_CAF1 40866 114 - 1 ACAGAAAAAUGCAACACGAUCGCAAGGUGCUAACGAAACGCAAAAGUCGUAAGGACAAAUCGGCCGCAGCAAUGGCAGCGGCAGCUGCUGCAGCGGUUCUGGAAGGGAAUGCGG .((((...........(((((....)((.((.((((..........)))).)).))..))))(((((.(((...(((((....)))))))).)))))))))............. ( -34.30) >DroMoj_CAF1 51426 114 - 1 GCAGAAAGUUGCAAAAUGAGCGCAAGGCUCUCACGAAGCGCAAGAGCCGCAAGGAGAAAUCGGCAGCAGCGAUGGCAGCCGCCGCUGCAGCUGCUGUGCUGGAAGGAAAUGCAG ((((....)))).........(((.((((((..((...))..))))))((...((....)).((((((((....(((((....))))).))))))))))..........))).. ( -43.60) >DroAna_CAF1 54650 114 - 1 CCAGAAAACUUCACAAUGAACGGAAAGCCCUCACAAAGCGGAAGAGUCGGAAGGAGAAGUCCGCUGCUGCGAUGGCGGCUGCCGCCGCUGCUGCGGUUCUGGAGGGGAAUGCCG ....................(((....((((..((.((((((....((....)).....))))))((((((..((((((....))))))..))))))..))..))))....))) ( -46.00) >consensus GCAGAAAAAUGCAAAAUGAACGCAAGGCGCUCACGAAGCGCAAAAGCCGCAAGGAGAAAUCGGCAGCAGCAAUGGCAGCCGCAGCUGCUGCCGCUGUGCUGGAAGGGAAUGCCG .(((..................................((......((....))......))(((((.((...((((((....)))))))).))))).)))............. (-24.00 = -23.50 + -0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:26 2006