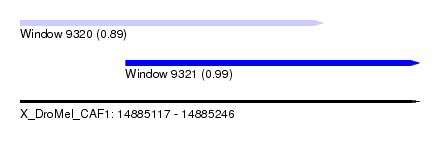
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,885,117 – 14,885,246 |
| Length | 129 |
| Max. P | 0.989759 |

| Location | 14,885,117 – 14,885,215 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 91.28 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -26.52 |
| Energy contribution | -25.80 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.887135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
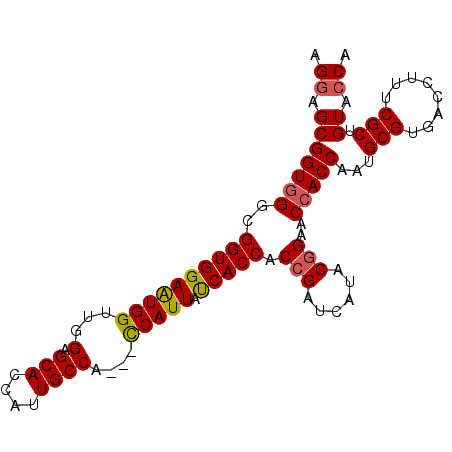
>X_DroMel_CAF1 14885117 98 + 22224390 ACCCACAAGAGACACACAGCUGAAUGAAGCUAAAAGGAGCGGUGGGCGGUGGAAUGGUUGGAGCACCAUUGCCA---CCAUUACCACCACCGAUCAUACAG ...................(((.((((.(((......)))(((((..(((((((((((......)))))).)))---))....))))).....)))).))) ( -32.30) >DroSec_CAF1 31183 97 + 1 ACCCACAAGAGACACAGAGCUGAAUGAAGCUAG-AGGAGCGGUGGGCGGUGGAAUGGUUGGAGCACCAUUGCCA---UCAUUAUCACCACCGAUCAUACGG ..((.............((((......))))..-..((.((((((..(((((((((((......)))))).)))---)).......)))))).))....)) ( -28.90) >DroSim_CAF1 22549 97 + 1 ACCCACAAGAGACACAGAGCUGAAUGAAGCUAG-AGAAGCGGUGGGCGGUGGAAUGGUUGGAGCACCAUUGCCA---UCAUUAUCACCACCGAUCAUACGG ..((.............((((......)))).(-(....((((((..(((((((((((......)))))).)))---)).......)))))).))....)) ( -28.40) >DroEre_CAF1 30215 97 + 1 ACCCACAAGAGACACAGAGCUGAAUGAAGCUAG-AGGAGCGGUUGGCGGUGGAGUGGUUGGAGCACCAUUGCCA---CCAUUACCACCACCGAUCAUACGG ..((........(....((((......))))..-..)...((((((.(((((((((((.((..........)))---))))).))))).))))))....)) ( -32.20) >DroYak_CAF1 30729 93 + 1 -------AGAGACAUAGAGCUGAAUGAAGCUAG-AGGAGCGGUGGGCGGUGGAGUGGUUGGAGCACCAUUGCCAUUACCAUUAUCACCACCGAUCAUACGG -------..........((((......))))..-..((.((((((..((((((((((((((....)))..)))))).)))))....)))))).))...... ( -30.80) >consensus ACCCACAAGAGACACAGAGCUGAAUGAAGCUAG_AGGAGCGGUGGGCGGUGGAAUGGUUGGAGCACCAUUGCCA___CCAUUAUCACCACCGAUCAUACGG .................((((......)))).........(((.((.((((((((((...(.(((....))))....))))).))))).)).)))...... (-26.52 = -25.80 + -0.72)

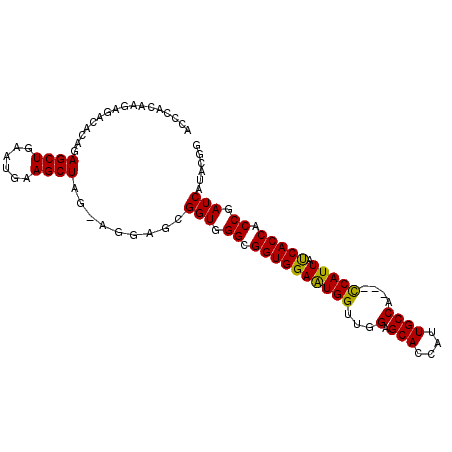
| Location | 14,885,151 – 14,885,246 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 95.63 |
| Mean single sequence MFE | -33.50 |
| Consensus MFE | -31.26 |
| Energy contribution | -31.14 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14885151 95 + 22224390 AGGAGCGGUGGGCGGUGGAAUGGUUGGAGCACCAUUGCCA---CCAUUACCACCACCGAUCAUACAGAACCACCAAUGCGUGACCUUUCGCUGUACCA .((.(((((((.((((((..(((((((.(((....)))..---)))..)))))))))).((.....)).)))))...(((........))).)).)). ( -34.10) >DroSec_CAF1 31216 95 + 1 AGGAGCGGUGGGCGGUGGAAUGGUUGGAGCACCAUUGCCA---UCAUUAUCACCACCGAUCAUACGGAACCACCAAUGCGUGACCUUUCGCUGUACCA .((.(((((((..(((((((((((......)))))).)))---))..........(((......)))..)))))...(((........))).)).)). ( -32.90) >DroSim_CAF1 22582 95 + 1 AGAAGCGGUGGGCGGUGGAAUGGUUGGAGCACCAUUGCCA---UCAUUAUCACCACCGAUCAUACGGAACCACCAAUGCGUGACCUUUCGCUGUACCA ......(((((..(((((((((((......)))))).)))---))..........(((......)))..)))))..(((((((....)))).)))... ( -29.60) >DroEre_CAF1 30248 95 + 1 AGGAGCGGUUGGCGGUGGAGUGGUUGGAGCACCAUUGCCA---CCAUUACCACCACCGAUCAUACGGAACCACCAAUGCGUGACCUUUCGCUGUACCA .((.((((((((.(((((((((((.((..........)))---))))).))))).))))))....((.....))...))((((....))))....)). ( -34.60) >DroYak_CAF1 30755 98 + 1 AGGAGCGGUGGGCGGUGGAGUGGUUGGAGCACCAUUGCCAUUACCAUUAUCACCACCGAUCAUACGGAACCACCAAUGCGUGACCUUUCGCUGUACCA .((.(((((((..(((((((((((..(.(((....))))...)))))).))))).(((......)))..)))))...(((........))).)).)). ( -36.30) >consensus AGGAGCGGUGGGCGGUGGAAUGGUUGGAGCACCAUUGCCA___CCAUUAUCACCACCGAUCAUACGGAACCACCAAUGCGUGACCUUUCGCUGUACCA .((.(((((((..((((((((((...(.(((....))))....))))).))))).(((......)))..)))))...(((........))).)).)). (-31.26 = -31.14 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:20 2006