| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,879,995 – 14,880,141 |
| Length | 146 |
| Max. P | 0.875517 |

| Location | 14,879,995 – 14,880,101 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 83.96 |
| Mean single sequence MFE | -24.57 |
| Consensus MFE | -15.52 |
| Energy contribution | -16.30 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14879995 106 + 22224390 UUUUGCA-CUGCAU---UGUACAGUCGAUGAAAUGUGCUACUUAUGAUUGGCACAAGUGAUUGUGGCCAAAAUAACGAUAGUGGCGUUAACAAUAUGCCAAAAUACAUGU ....(((-(..(((---((......)))))....))))....((((.(((((((((....)))).)))))...........((((((.......)))))).....)))). ( -29.00) >DroSec_CAF1 26310 98 + 1 AU--GCAAUUGCAUAAAUGCA-AGUCCAUGAAAUGUGCUAACUAUUAUUGGCACAUGUGAUUGUGGCCAAAAUAACGAU---------AACGAUAUGCCAAAAUACAUGU ..--....(((((....))))-)...((((..(((((((((......)))))))))...(((.((((....(((.((..---------..)).))))))).))).)))). ( -21.90) >DroSim_CAF1 17715 98 + 1 AU--GCAAUUGCAUAAAUGCA-AGUCCAUGAAAUGUGCUAAUUAUCAUUGGCACAUGUGAUUGUGGCCAAAAUAACGAU---------AACGAUAUGCCAAAAUACAUGU ..--....(((((....))))-)...((((..((((((((((....))))))))))...(((.((((....(((.((..---------..)).))))))).))).)))). ( -22.80) >consensus AU__GCAAUUGCAUAAAUGCA_AGUCCAUGAAAUGUGCUAAUUAUCAUUGGCACAUGUGAUUGUGGCCAAAAUAACGAU_________AACGAUAUGCCAAAAUACAUGU ....(((..((((....))))..(.(((..(.(((((((((......)))))))))....)..))).)...........................)))............ (-15.52 = -16.30 + 0.78)

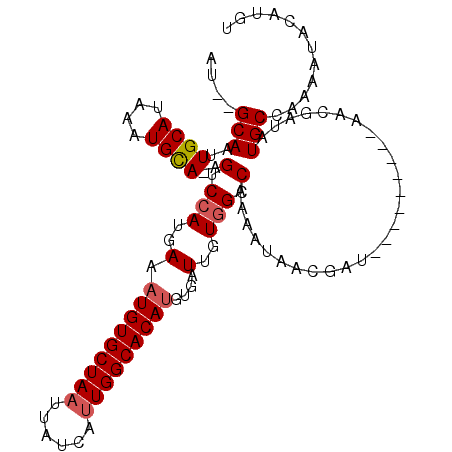

| Location | 14,880,021 – 14,880,141 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.71 |
| Mean single sequence MFE | -24.77 |
| Consensus MFE | -18.63 |
| Energy contribution | -19.13 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.754787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14880021 120 + 22224390 AAAUGUGCUACUUAUGAUUGGCACAAGUGAUUGUGGCCAAAAUAACGAUAGUGGCGUUAACAAUAUGCCAAAAUACAUGUAGCAUACACACGUGUACAUACAUUCAAUUUGAAAGCAUUG .((((((((((..(((.(((((((((....)))).)))))...........((((((.......)))))).....)))))))).((((....))))..................))))). ( -30.20) >DroSec_CAF1 26337 103 + 1 AAAUGUGCUAACUAUUAUUGGCACAUGUGAUUGUGGCCAAAAUAACGAU---------AACGAUAUGCCAAAAUACAUGUAGCA--------UGUACAUACAUUCAUUUUGAAAGCAUUG ..(((((((((......)))))))))((((.((((((....(((.((..---------..)).))))))....((((((...))--------))))...))).))))............. ( -21.60) >DroSim_CAF1 17742 102 + 1 AAAUGUGCUAAUUAUCAUUGGCACAUGUGAUUGUGGCCAAAAUAACGAU---------AACGAUAUGCCAAAAUACAUGUAGCA--------UGUACAUACAUUCAUUUUGAAAGCAUU- ..((((((((((....))))))))))((((.((((((....(((.((..---------..)).))))))....((((((...))--------))))...))).))))............- ( -22.50) >consensus AAAUGUGCUAAUUAUCAUUGGCACAUGUGAUUGUGGCCAAAAUAACGAU_________AACGAUAUGCCAAAAUACAUGUAGCA________UGUACAUACAUUCAUUUUGAAAGCAUUG ....(((((........((((((((......))).)))))............................((((((..(((((((((......))))...)))))..))))))..))))).. (-18.63 = -19.13 + 0.50)

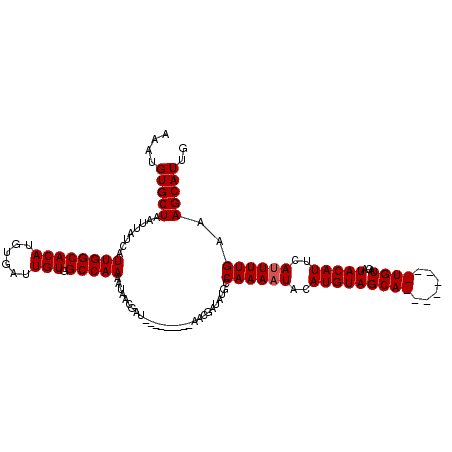

| Location | 14,880,021 – 14,880,141 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.71 |
| Mean single sequence MFE | -26.07 |
| Consensus MFE | -19.23 |
| Energy contribution | -19.57 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14880021 120 - 22224390 CAAUGCUUUCAAAUUGAAUGUAUGUACACGUGUGUAUGCUACAUGUAUUUUGGCAUAUUGUUAACGCCACUAUCGUUAUUUUGGCCACAAUCACUUGUGCCAAUCAUAAGUAGCACAUUU ..((((.(((.....))).))))(((((....)))))(((((.(((...((((((((.((.....((((..((....))..))))......))..))))))))..))).)))))...... ( -29.60) >DroSec_CAF1 26337 103 - 1 CAAUGCUUUCAAAAUGAAUGUAUGUACA--------UGCUACAUGUAUUUUGGCAUAUCGUU---------AUCGUUAUUUUGGCCACAAUCACAUGUGCCAAUAAUAGUUAGCACAUUU ...((((..(..(((((((((..(((((--------((...)))))))....)))).)))))---------.....(((((((((.(((......)))))))).)))))..))))..... ( -24.30) >DroSim_CAF1 17742 102 - 1 -AAUGCUUUCAAAAUGAAUGUAUGUACA--------UGCUACAUGUAUUUUGGCAUAUCGUU---------AUCGUUAUUUUGGCCACAAUCACAUGUGCCAAUGAUAAUUAGCACAUUU -..((((.....(((((((((..(((((--------((...)))))))....)))).)))))---------...(((((((((((.(((......)))))))).)))))).))))..... ( -24.30) >consensus CAAUGCUUUCAAAAUGAAUGUAUGUACA________UGCUACAUGUAUUUUGGCAUAUCGUU_________AUCGUUAUUUUGGCCACAAUCACAUGUGCCAAUAAUAAUUAGCACAUUU ...((((.((((((((.(((((.(((..........)))))))).))))))))......................((((((((((.(((......)))))))).)))))..))))..... (-19.23 = -19.57 + 0.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:19 2006