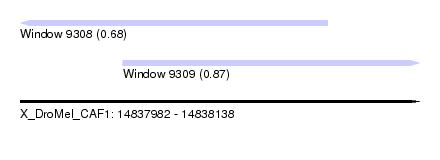
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,837,982 – 14,838,138 |
| Length | 156 |
| Max. P | 0.865600 |

| Location | 14,837,982 – 14,838,102 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.78 |
| Mean single sequence MFE | -59.60 |
| Consensus MFE | -50.06 |
| Energy contribution | -52.15 |
| Covariance contribution | 2.09 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14837982 120 - 22224390 GCUGCACCCUCGAUGACCACGCGACUGUCGCGACUUGGCGGCGAGGAGUCCACCUCGCCACCCUCGUCGAUGGUGUCCUGGCGGGCAAGGGUGGCCUGGCUAGCUUGCAGCUUGGGCGCC ((((((((.(((((((...((((.....))))....((.(((((((......))))))).)).))))))).)))))...))).......((((.((.((((.(....))))).)).)))) ( -57.50) >DroVir_CAF1 4351 120 - 1 GCGCCGCCAUCGAUGACCACCCGGCUGUCACGGCUGGGCGGCGAUGAGUCAACCUCGCCGCCCUCGUCGAUGGUGUCCUGGCGGGCCAGCGUCGCCUGGCUGGCCUGCAGCUUGGGCGCC ((((((((((((((((......(((((...)))))(((((((((.(......).))))))))))))))))))))......((((((((((........))))))))))......))))). ( -73.50) >DroGri_CAF1 5076 120 - 1 GCAGCGCCAUCGAUGACCACCCGACUGUCCCGACUAGGUGGCGAGGAGUCAACCUCGCCGCCCUCAUCGAUGGUGUCCUGGCGGGCGAGUGUUGCCUGGCUAGCUUGUAGCUUGGGCGCU ..((((((((((((((.....((.......))....((((((((((......)))))))))).)))))))))))((((.(((..((((((...((...))..)))))).))).))))))) ( -59.20) >DroWil_CAF1 5977 120 - 1 CCUCCGCCCUCCAUGACCACACGACUGUCACGACUGGGCGGGGAGGAGUCAACCUCGCCGCCCUCAUCGAUGGUAUCCUGGCGUGCUAGGGUAGCUUGGCUAGCCUGCAUUUUGGGUGCC ....(((((.....(.(((....((((((......((((((.((((......)))).)))))).....))))))....))))((((.(((.((((...)))).)))))))...))))).. ( -50.80) >DroMoj_CAF1 4301 120 - 1 GCACCGCCAUCGAUGACCACACGACUGUCCCGGCUGGGUGGCGACGAGUCCACCUCGCCGCCCUCGUCGAUGGUGUCCUGACGAGCGAGCGUCGCCUGGCUAGCCUGCAGCUUGGGCGCC ((..((((((((((((.....((.......))...(((((((((.(......).)))))))))))))))))))))....((((......))))((((((((.(....)))).))))))). ( -52.00) >DroPer_CAF1 4843 120 - 1 GCCCCUCCAUCGAUGACUAUACGGCUGUCGCGACUGGGUGGAGAGGAGUCCACCUCGCCGCCCUCGUCGAUGGUGUCCUGGCGGGCGAGCGUCGCCUGGCUCGCCUGCAGCUUGGGCGCC (((((.((((((((((.......((....))....((((((.((((......)))).)))))))))))))))).).....((((((((((........)))))))))).....))))... ( -64.60) >consensus GCACCGCCAUCGAUGACCACACGACUGUCACGACUGGGCGGCGAGGAGUCAACCUCGCCGCCCUCGUCGAUGGUGUCCUGGCGGGCGAGCGUCGCCUGGCUAGCCUGCAGCUUGGGCGCC ((...(((((((((((.....((.......))...(((((((((((......))))))))))))))))))))))((((..((((((((((........)))))))))).....)))))). (-50.06 = -52.15 + 2.09)


| Location | 14,838,022 – 14,838,138 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.23 |
| Mean single sequence MFE | -52.59 |
| Consensus MFE | -30.32 |
| Energy contribution | -30.85 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14838022 116 + 22224390 CAGGACACCAUCGACGAGGGUGGCGAGGUGGACUCCUCGCCGCCAAGUCGCGACAGUCGCGUGGUCAUCGAGGGUGCAGCCGUCUCCUCGGCCACCGUGG-GACCAUCGCCGCCAGU ..((.(......(((...((((((((((......))))))))))..)))((((..(((.((((((..((((((..((....))..))))))..)))))).-)))..)))).)))... ( -57.30) >DroEre_CAF1 4082 110 + 1 CAGGACACCAUCGACGAGGGUGGCGAGGUGGACUCCUCGCCGCCAAGUCGCGACAGUCGCGUGGUCAUCGAGGGUGCAGC------CUCGGCCACCGUGG-GACCAUCGCCGCCAGU ..((.(......(((...((((((((((......))))))))))..)))((((..(((.((((((..((((((......)------)))))..)))))).-)))..)))).)))... ( -55.00) >DroWil_CAF1 6017 114 + 1 CAGGAUACCAUCGAUGAGGGCGGCGAGGUUGACUCCUCCCCGCCCAGUCGUGACAGUCGUGUGGUCAUGGAGGGCGGAGGAACCAACACAACCAACGCAGUCACCAUUGCCUCU--- .(((.......(((((..(((.(((.(((((.....(((((((((..(((((((.........))))))).)))))).))).......)))))..))).)))..))))))))..--- ( -43.11) >DroYak_CAF1 4111 116 + 1 CAGGACACCAUCGACGAGGGUGGCGAGGUGGACUCCUCGCCGCCAAGUCGCGACAGUCGCGUGGUUAUCGAGGGUGCAGCCGUCACCUCGGCCACCGUGG-GACCAUCGCCGCCAGU ..((.(......(((...((((((((((......))))))))))..)))((((..(((.((((((..((((((..((....))..))))))..)))))).-)))..)))).)))... ( -56.60) >DroMoj_CAF1 4341 104 + 1 CAGGACACCAUCGACGAGGGCGGCGAGGUGGACUCGUCGCCACCCAGCCGGGACAGUCGUGUGGUCAUCGAUGGCGGUGCAGCCGCC---------ACA----GCCACGUCCACAGC ..(((((((((((((...(((((((((.....))))))))).(((....)))...)))).)))))....(.((((((.....)))))---------)).----.....))))..... ( -52.20) >DroAna_CAF1 4208 105 + 1 CAGGACACCAUCGACGAGGGCGGCGAGGUGGACUCCUCCCCGCCCAGUCGCGACAGUCGUGUGGUCAUCGAAGGUGCCACAGGAGCAGCAGGC--GG----------UGGUGGCAGU .....((((((((((..((((((.((((......)))).)))))).)))((..(..((.(((((.(((.....)))))))).)))..))....--))----------)))))..... ( -51.30) >consensus CAGGACACCAUCGACGAGGGCGGCGAGGUGGACUCCUCGCCGCCAAGUCGCGACAGUCGCGUGGUCAUCGAGGGUGCAGCAGCCACCUCGGCCACCGUGG_GACCAUCGCCGCCAGU .....((((.((((.((.((((((((((......))))))))))....((((.....))))...)).)))).))))......................................... (-30.32 = -30.85 + 0.53)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:10 2006