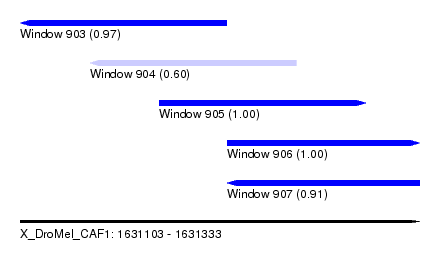
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,631,103 – 1,631,333 |
| Length | 230 |
| Max. P | 0.999894 |

| Location | 1,631,103 – 1,631,222 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 91.25 |
| Mean single sequence MFE | -31.68 |
| Consensus MFE | -27.74 |
| Energy contribution | -28.38 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1631103 119 - 22224390 ACACCCACACUCUCGUGAAUUGGCCAAUCGGGCCCAUAGCCGUCGUAGCCAUCGACUGGCUGGCCAAAAGUGCACUACACACAGUAUACAACUUAUGCAGGCACCUUACUAUCUGCUCU .....(((......)))..(((((((..(((((.....)))((((.......))))))..)))))))..((((.((.((...(((.....)))..)).))))))............... ( -30.20) >DroSec_CAF1 26235 118 - 1 ACACCCACGCUCUCGUGAAUUGGCCAAUCGGACCCAU-GCCGUCGUAGCCAUCGACUGGCUGGCCAAAAGUGUACUACACACAGUAUACAACUUAUGCAGGCACCUUACUAUCUGCUCU .....((((....))))..(((((((...(....)..-(((((((.......)))).))))))))))..(((((((......))))))).......((((..(......)..))))... ( -34.10) >DroSim_CAF1 42760 118 - 1 ACACCCACGCUCUCGUGAAUUGGCCAAUCGGACCCAU-GCCGUCGUAGCCAUCGACUGGCUGGCCAAAAGUGUACUACACACAGUAUACAACUUAUGCAGGCACCUUACUAUCUGCUCU .....((((....))))..(((((((...(....)..-(((((((.......)))).))))))))))..(((((((......))))))).......((((..(......)..))))... ( -34.10) >DroEre_CAF1 27368 109 - 1 ACACCCACGCUCUCGUGAAUUGGCCAAUCGGGCCCAUAGCCAU---------CGACUGGCUGGCCAAAAGUGCACUACACACAGUAUACAGCUUAUGCAGGCACCUUACUAUCUGCUC- .....((((....))))....((((.....))))..((((((.---------....))))))(((....(((.(((......))).))).((....)).)))................- ( -26.80) >DroYak_CAF1 30572 118 - 1 ACACCCACGCUCUCGUGAAUUGACCAAUCGGGCCCAAAGCCAUCGCUCGCAUCGGCUGGCUGGCCAAAAGUGCACUACACACAGUGUACAGCUUAUGCAGGCACCUUACUAUCUGCUC- ........((....((((..((.((.....((((...(((((..(((......))))))))))))....(((((((......))))))).((....)).))))..)))).....))..- ( -33.20) >consensus ACACCCACGCUCUCGUGAAUUGGCCAAUCGGGCCCAUAGCCGUCGUAGCCAUCGACUGGCUGGCCAAAAGUGCACUACACACAGUAUACAACUUAUGCAGGCACCUUACUAUCUGCUCU .....((((....))))..(((((((...(....)...(((((((.......))).)))))))))))..(((((((......))))))).......((((..(......)..))))... (-27.74 = -28.38 + 0.64)

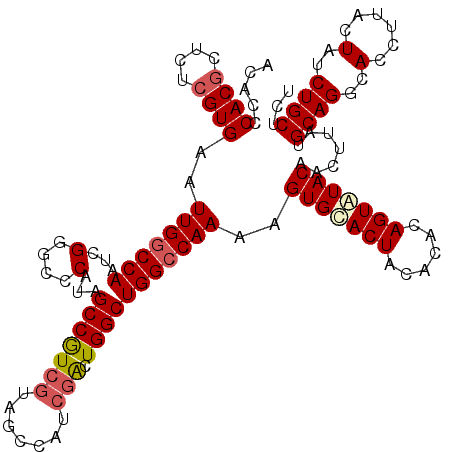
| Location | 1,631,143 – 1,631,262 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 91.76 |
| Mean single sequence MFE | -31.84 |
| Consensus MFE | -24.50 |
| Energy contribution | -24.66 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1631143 119 - 22224390 UACUUGUAUGCCCAAGUUACACGAUUCUAUCGAUUUUCGUACACCCACACUCUCGUGAAUUGGCCAAUCGGGCCCAUAGCCGUCGUAGCCAUCGACUGGCUGGCCAAAAGUGCACUACA .(((((......)))))....((((...))))......((((...(((......)))..(((((((..(((((.....)))((((.......))))))..)))))))..))))...... ( -32.40) >DroSec_CAF1 26275 118 - 1 UACUUGUAUGCCCAAGUUACACGAUUCUAUCGAUUUUCGUACACCCACGCUCUCGUGAAUUGGCCAAUCGGACCCAU-GCCGUCGUAGCCAUCGACUGGCUGGCCAAAAGUGUACUACA .(((((......)))))....((((...))))......((((((.((((....))))..(((((((...(....)..-(((((((.......)))).))))))))))..)))))).... ( -34.80) >DroSim_CAF1 42800 118 - 1 UACUUGUAUGCCCAAGUUACACGAUUCUAUCGAUUUUCGUACACCCACGCUCUCGUGAAUUGGCCAAUCGGACCCAU-GCCGUCGUAGCCAUCGACUGGCUGGCCAAAAGUGUACUACA .(((((......)))))....((((...))))......((((((.((((....))))..(((((((...(....)..-(((((((.......)))).))))))))))..)))))).... ( -34.80) >DroEre_CAF1 27407 110 - 1 UACUUGUAUGCCUAAGUUACACGAUUCUAUCGAUUUUCGUACACCCACGCUCUCGUGAAUUGGCCAAUCGGGCCCAUAGCCAU---------CGACUGGCUGGCCAAAAGUGCACUACA (((((....(((.........((((((...(((....(((......)))...))).))))))((((.((((((.....)))..---------))).)))).)))...)))))....... ( -28.10) >DroYak_CAF1 30611 119 - 1 UACUUGUAUGCCUAAGUUACACGAUUCUAUUGAUUUUCGUACACCCACGCUCUCGUGAAUUGACCAAUCGGGCCCAAAGCCAUCGCUCGCAUCGGCUGGCUGGCCAAAAGUGCACUACA ....(((((((((..((((.((((.((....))...)))).....((((....))))...))))......((((...(((((..(((......))))))))))))...)).))).)))) ( -29.10) >consensus UACUUGUAUGCCCAAGUUACACGAUUCUAUCGAUUUUCGUACACCCACGCUCUCGUGAAUUGGCCAAUCGGGCCCAUAGCCGUCGUAGCCAUCGACUGGCUGGCCAAAAGUGCACUACA .(((((......)))))...((((.((....))...)))).(((.((((....))))..(((((((...(....)...(((((((.......))).)))))))))))..)))....... (-24.50 = -24.66 + 0.16)

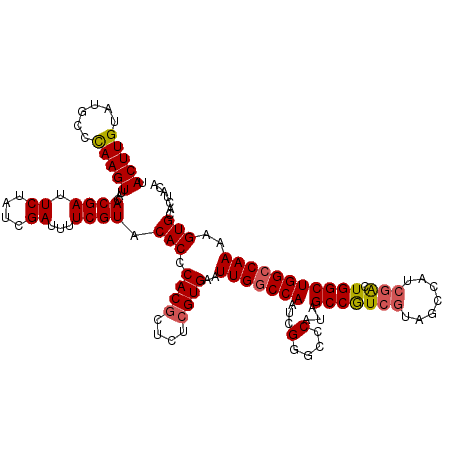
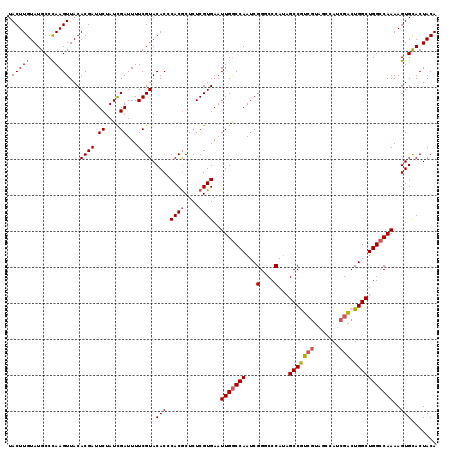
| Location | 1,631,183 – 1,631,302 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 92.90 |
| Mean single sequence MFE | -34.04 |
| Consensus MFE | -33.74 |
| Energy contribution | -33.50 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.54 |
| SVM RNA-class probability | 0.999366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1631183 119 + 22224390 CUAUGGGCCCGAUUGGCCAAUUCACGAGAGUGUGGGUGUACGAAAAUCGAUAGAAUCGUGUAACUUGGGCAUACAAGUACGUAUGUUUAAUGUUGCAUGCAUGUGGAAUGUACAUACAU .....((((.....))))....(((....)))((..((((((....((.(((....((((((((((((((((((......)))))))))).))))))))..))).)).))))))..)). ( -39.10) >DroSec_CAF1 26315 118 + 1 C-AUGGGUCCGAUUGGCCAAUUCACGAGAGCGUGGGUGUACGAAAAUCGAUAGAAUCGUGUAACUUGGGCAUACAAGUACGUAUGUUUAAUGUUGCAUGCAUGUGGAAUAUACAUACAU (-(((....(((((..(..(((((((....)))))))....)..))))).......((((((((((((((((((......)))))))))).))))))))))))................ ( -33.00) >DroSim_CAF1 42840 118 + 1 C-AUGGGUCCGAUUGGCCAAUUCACGAGAGCGUGGGUGUACGAAAAUCGAUAGAAUCGUGUAACUUGGGCAUACAAGUACGUAUGUUUAAUGUUGCAUGCAUGUGGAAUAUACAUACAU (-(((....(((((..(..(((((((....)))))))....)..))))).......((((((((((((((((((......)))))))))).))))))))))))................ ( -33.00) >DroEre_CAF1 27438 113 + 1 CUAUGGGCCCGAUUGGCCAAUUCACGAGAGCGUGGGUGUACGAAAAUCGAUAGAAUCGUGUAACUUAGGCAUACAAGUACGUAUGUUUAAUGUUGCAUGCAUGUGGAAUGUAU------ .....((((.....)))).(((((((....)))))))(((((....((.(((....((((((((((((((((((......)))))))))).))))))))..))).)).)))))------ ( -36.10) >DroYak_CAF1 30651 113 + 1 CUUUGGGCCCGAUUGGUCAAUUCACGAGAGCGUGGGUGUACGAAAAUCAAUAGAAUCGUGUAACUUAGGCAUACAAGUACGUAUGUUAAAUGUUGCAUGCAUGUGGAAUGUAC------ .....((((.....)))).(((((((....)))))))(((((....((.(((....((((((((((..((((((......))))))..)).))))))))..))).)).)))))------ ( -29.00) >consensus CUAUGGGCCCGAUUGGCCAAUUCACGAGAGCGUGGGUGUACGAAAAUCGAUAGAAUCGUGUAACUUGGGCAUACAAGUACGUAUGUUUAAUGUUGCAUGCAUGUGGAAUGUACAUACAU .....((((.....)))).(((((((....)))))))(((((....((.(((....((((((((((((((((((......)))))))))).))))))))..))).)).)))))...... (-33.74 = -33.50 + -0.24)

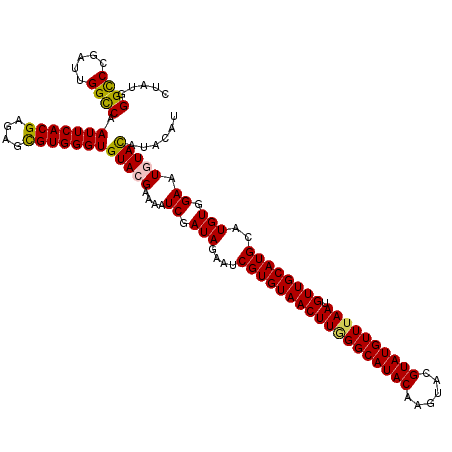
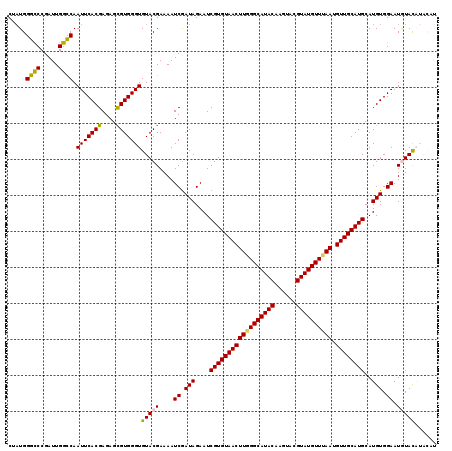
| Location | 1,631,222 – 1,631,333 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -26.70 |
| Energy contribution | -26.94 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.55 |
| Structure conservation index | 0.87 |
| SVM decision value | 4.42 |
| SVM RNA-class probability | 0.999894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1631222 111 + 22224390 ACGAAAAUCGAUAGAAUCGUGUAACUUGGGCAUACAAGUACGUAUGUUUAAUGUUGCAUGCAUGUGGAAUGUACAUACAUAUAUACGCAUGCCUAUAAUCAUGCUUAUGAG .((.....))((((....(((((((((((((((((......)))))))))).)))))))(((((((..(((((......))))).)))))))))))..((((....)))). ( -31.40) >DroSec_CAF1 26353 111 + 1 ACGAAAAUCGAUAGAAUCGUGUAACUUGGGCAUACAAGUACGUAUGUUUAAUGUUGCAUGCAUGUGGAAUAUACAUACAUAUAUACGCAUGCCUAUAAUCAUGCAUAUGAG .((.....))((((....(((((((((((((((((......)))))))))).)))))))(((((((..(((((......))))).)))))))))))..((((....)))). ( -31.40) >DroSim_CAF1 42878 111 + 1 ACGAAAAUCGAUAGAAUCGUGUAACUUGGGCAUACAAGUACGUAUGUUUAAUGUUGCAUGCAUGUGGAAUAUACAUACAUAUAUACGCAUGCCUAUAAUCAUGCAUAUGAG .((.....))((((....(((((((((((((((((......)))))))))).)))))))(((((((..(((((......))))).)))))))))))..((((....)))). ( -31.40) >DroEre_CAF1 27477 103 + 1 ACGAAAAUCGAUAGAAUCGUGUAACUUAGGCAUACAAGUACGUAUGUUUAAUGUUGCAUGCAUGUGGAAUGUAU--------GUAUGCAUGCCAAUAAUCAUGCAUAUAAG .......((.(((....((((((((((((((((((......)))))))))).))))))))..))).)).....(--------(((((((((........)))))))))).. ( -33.30) >DroYak_CAF1 30690 103 + 1 ACGAAAAUCAAUAGAAUCGUGUAACUUAGGCAUACAAGUACGUAUGUUAAAUGUUGCAUGCAUGUGGAAUGUAC--------GUACGCAUGCCAAUAAUCAUGCAUAUAAG ((((...((....)).))))(((((((..((((((......))))))..)).)))))((((((((((.((((..--------....)))).))).....)))))))..... ( -26.00) >consensus ACGAAAAUCGAUAGAAUCGUGUAACUUGGGCAUACAAGUACGUAUGUUUAAUGUUGCAUGCAUGUGGAAUGUACAUACAUAUAUACGCAUGCCUAUAAUCAUGCAUAUGAG .......((.(((....((((((((((((((((((......)))))))))).))))))))..))).)).............((((.(((((........))))).)))).. (-26.70 = -26.94 + 0.24)

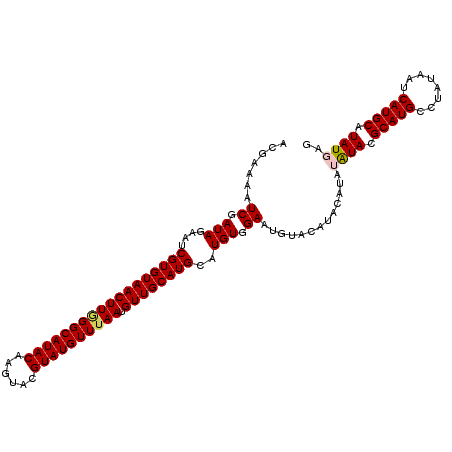
| Location | 1,631,222 – 1,631,333 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -23.09 |
| Consensus MFE | -17.81 |
| Energy contribution | -17.89 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1631222 111 - 22224390 CUCAUAAGCAUGAUUAUAGGCAUGCGUAUAUAUGUAUGUACAUUCCACAUGCAUGCAACAUUAAACAUACGUACUUGUAUGCCCAAGUUACACGAUUCUAUCGAUUUUCGU .......(((((........)))))(((..(((((((((.......)))))))))..........((((((....)))))).......)))((((.((....))...)))) ( -22.10) >DroSec_CAF1 26353 111 - 1 CUCAUAUGCAUGAUUAUAGGCAUGCGUAUAUAUGUAUGUAUAUUCCACAUGCAUGCAACAUUAAACAUACGUACUUGUAUGCCCAAGUUACACGAUUCUAUCGAUUUUCGU ...(((((((((........)))))))))...(((((((((.......))))))))).............(((((((......)))).)))((((.((....))...)))) ( -25.40) >DroSim_CAF1 42878 111 - 1 CUCAUAUGCAUGAUUAUAGGCAUGCGUAUAUAUGUAUGUAUAUUCCACAUGCAUGCAACAUUAAACAUACGUACUUGUAUGCCCAAGUUACACGAUUCUAUCGAUUUUCGU ...(((((((((........)))))))))...(((((((((.......))))))))).............(((((((......)))).)))((((.((....))...)))) ( -25.40) >DroEre_CAF1 27477 103 - 1 CUUAUAUGCAUGAUUAUUGGCAUGCAUAC--------AUACAUUCCACAUGCAUGCAACAUUAAACAUACGUACUUGUAUGCCUAAGUUACACGAUUCUAUCGAUUUUCGU .........((((..((((((((((((..--------...........))))))))(((.(((..((((((....))))))..))))))............))))..)))) ( -21.32) >DroYak_CAF1 30690 103 - 1 CUUAUAUGCAUGAUUAUUGGCAUGCGUAC--------GUACAUUCCACAUGCAUGCAACAUUUAACAUACGUACUUGUAUGCCUAAGUUACACGAUUCUAUUGAUUUUCGU ..........(((((...(((((((((((--------(((.........((....))..........)))))))..)))))))..))))).((((.((....))...)))) ( -21.21) >consensus CUCAUAUGCAUGAUUAUAGGCAUGCGUAUAUAUGUAUGUACAUUCCACAUGCAUGCAACAUUAAACAUACGUACUUGUAUGCCCAAGUUACACGAUUCUAUCGAUUUUCGU ....((((((((........))))))))......................((((((((................)))))))).........((((.((....))...)))) (-17.81 = -17.89 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:09 2006