
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,813,341 – 14,813,483 |
| Length | 142 |
| Max. P | 0.959978 |

| Location | 14,813,341 – 14,813,443 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 81.61 |
| Mean single sequence MFE | -31.87 |
| Consensus MFE | -17.28 |
| Energy contribution | -17.53 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14813341 102 + 22224390 AGUGUGACAGUGUGCGUGUGCGUCUAAAUGUGUGUGUUUGUGAGAGUUGGUCAACCCACUAACACUUGCACACACUCGCA--CAAAAAGAAAGUGCCAAGAAUU .........(.(..(.((((((.......((((((((..(((..(((.((....)).)))..)))..)))))))).))))--))........)..))....... ( -31.80) >DroSec_CAF1 7352 104 + 1 AGUGUGCCAGUGUGCGUGUGCGUCUGAAUUUGUGUGUUUGUGAGUGUUGGUCAACCCACUAACACUUGCACACACUCGCACACAAAAAGAAAGUGCCAAGAAUU .((((((.((((((.(..((((........))))..).((..(((((((((......)))))))))..)))))))).))))))..................... ( -36.60) >DroYak_CAF1 141 78 + 1 AGUGUG----------UG--------------UGUGUUUGUGAGAGUUGGUCAACCCACUAACACUUGCACACACUCGCA--CAAAAAGAAAGUGCCAAGAAUU .(((.(----------((--------------(((((..(((..(((.((....)).)))..)))..)))))))).))).--...................... ( -27.20) >consensus AGUGUG_CAGUGUGCGUGUGCGUCU_AAU_UGUGUGUUUGUGAGAGUUGGUCAACCCACUAACACUUGCACACACUCGCA__CAAAAAGAAAGUGCCAAGAAUU .(((.........(((....)))..........((((.((..((.((((((......)))))).))..)).)))).)))......................... (-17.28 = -17.53 + 0.25)



| Location | 14,813,341 – 14,813,443 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 81.61 |
| Mean single sequence MFE | -30.17 |
| Consensus MFE | -17.52 |
| Energy contribution | -17.69 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.959978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14813341 102 - 22224390 AAUUCUUGGCACUUUCUUUUUG--UGCGAGUGUGUGCAAGUGUUAGUGGGUUGACCAACUCUCACAAACACACACAUUUAGACGCACACGCACACUGUCACACU ........((((.........)--)))..((((((((..(((((.(((((..........))))).)))))...(.....)..))))))))............. ( -28.80) >DroSec_CAF1 7352 104 - 1 AAUUCUUGGCACUUUCUUUUUGUGUGCGAGUGUGUGCAAGUGUUAGUGGGUUGACCAACACUCACAAACACACAAAUUCAGACGCACACGCACACUGGCACACU .....................((((((.((((((((...(((((.((((((........)))))).))))).........(.....).)))))))).)))))). ( -35.30) >DroYak_CAF1 141 78 - 1 AAUUCUUGGCACUUUCUUUUUG--UGCGAGUGUGUGCAAGUGUUAGUGGGUUGACCAACUCUCACAAACACA--------------CA----------CACACU ........((((.........)--))).((((((((...(((((.(((((..........))))).))))).--------------))----------)))))) ( -26.40) >consensus AAUUCUUGGCACUUUCUUUUUG__UGCGAGUGUGUGCAAGUGUUAGUGGGUUGACCAACUCUCACAAACACACA_AUU_AGACGCACACGCACACUG_CACACU ........(((.............))).((((((.....(((((.(((((..........))))).)))))............((....)).......)))))) (-17.52 = -17.69 + 0.17)

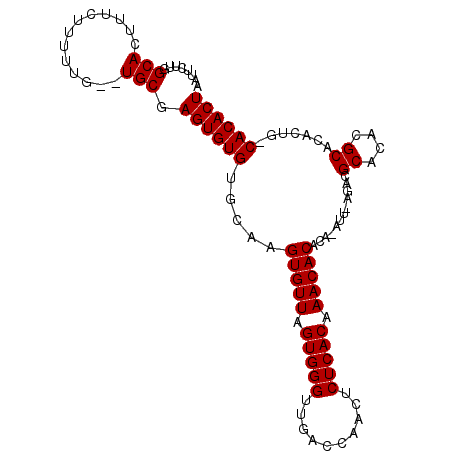
| Location | 14,813,367 – 14,813,483 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 91.76 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -24.97 |
| Energy contribution | -25.30 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14813367 116 + 22224390 AAUGUGUGUGUUUGUGAGAGUUGGUCAACCCACUAACACUUGCACACACUCGCA--CAAAAAGAAAGUGCCAAGAAUUGGCAAGCAAUCGGUAGUUAAUUCAAUAUGGCCGUAAAACG ...((((((((..(((..(((.((....)).)))..)))..)))))))).....--..........(.((((.((((((((.(.(....).).))))))))....)))))........ ( -33.60) >DroSec_CAF1 7378 118 + 1 AAUUUGUGUGUUUGUGAGUGUUGGUCAACCCACUAACACUUGCACACACUCGCACACAAAAAGAAAGUGCCAAGAAUUGGCAAGCAAUCGGUAGUUAAUUCAAUAUGGCCGUAAAACG ..(((((((((.((..(((((((((......)))))))))..)).......)))))))))......(.((((.((((((((.(.(....).).))))))))....)))))........ ( -35.70) >DroYak_CAF1 149 110 + 1 ------UGUGUUUGUGAGAGUUGGUCAACCCACUAACACUUGCACACACUCGCA--CAAAAAGAAAGUGCCAAGAAUUGGCAAGCAAUCGUUAGUUAAUUCAAUAUCGCAUUAAAGCG ------.((((.((..((.((((((......)))))).))..)).))))..(((--(.........))))...((((((((.(((....))).)))))))).....(((......))) ( -31.80) >consensus AAU_UGUGUGUUUGUGAGAGUUGGUCAACCCACUAACACUUGCACACACUCGCA__CAAAAAGAAAGUGCCAAGAAUUGGCAAGCAAUCGGUAGUUAAUUCAAUAUGGCCGUAAAACG .......((((.((..((.((((((......)))))).))..)).))))...................((((.((((((((.(.(....).).))))))))....))))......... (-24.97 = -25.30 + 0.33)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:54 2006