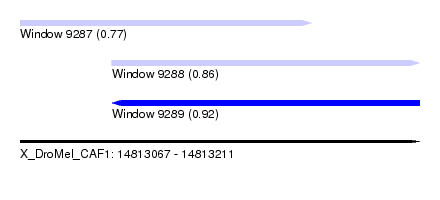
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,813,067 – 14,813,211 |
| Length | 144 |
| Max. P | 0.917157 |

| Location | 14,813,067 – 14,813,172 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.74 |
| Mean single sequence MFE | -36.50 |
| Consensus MFE | -23.33 |
| Energy contribution | -24.85 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14813067 105 + 22224390 ------GUGUGCAACUAAAACUCGCUCUUGCACUUC-CAAACU----GGGGAAAAGUUGGCUGCUUUUCCCGGCCACUUUGUUGCUGGU--U-UGGACCGAGUUGAA-AGUUUACGCAAU ------(((((.((((..(((((((....))...((-(((((.----.(.(((((((.(((((.......)))))))))).)).)..))--)-))))..)))))...-)))))))))... ( -40.70) >DroSec_CAF1 7015 106 + 1 ------GUGUGGAACUAAAACUCGCUCUUGCACUUC-CAAACU----GGGGAAAAGUUGGCUGCUUUUCCCGGCCACUUUGUUGCUGGU--UUUGGACCGAGUUGAA-AGUUUACGCAAU ------(((((((.....(((((((....))...((-((((..----.(((((((((.....))))))))).(((((......).))))--))))))..)))))...-..)))))))... ( -37.20) >DroSim_CAF1 6242 106 + 1 ------GUGUGGAACUAAAACUCGCUCUUGCACUUC-CAAACU----GGGGAAAAGUUGGCUGCUUUUCCCGGCCACUUUGUUGCUGGU--UUUGGACCGAGUUGAA-AGUUUACGCAAU ------(((((((.....(((((((....))...((-((((..----.(((((((((.....))))))))).(((((......).))))--))))))..)))))...-..)))))))... ( -37.20) >DroEre_CAF1 12259 111 + 1 GUUUGUGUGUGGAACUAAAACUCGCUCUUGCACUUC-CAAACU----GGGGAAAAGUUGGCUGCUUUUCCCGGCCACUUUGUUGCUGCU--U-UGGUCCGAGUUGAA-AGUUUACGCAAU .....((((((((.....((((((..((.(((.(..-((((.(----((((((((((.....))))))))...))).))))..).))).--.-.))..))))))...-..)))))))).. ( -36.10) >DroAna_CAF1 35150 113 + 1 ------GAGU-AAACUAAAACUCGCUGAUGCACUAGACAAGCGGGAGGGGGUAAAGGUGGUUGAUUUUCCCGGCGACUCUGUUGUUGGGCCUAUGGAUCCAGAUGAAUAGUUUAAGCAAU ------(..(-((((((..((((.((..(((.........)))..)).)))).....(((((.((...(((((((((...)))))))))...)).)).)))......)))))))..)... ( -31.30) >consensus ______GUGUGGAACUAAAACUCGCUCUUGCACUUC_CAAACU____GGGGAAAAGUUGGCUGCUUUUCCCGGCCACUUUGUUGCUGGU__U_UGGACCGAGUUGAA_AGUUUACGCAAU ......((((.(((((..(((((((....)).................(((((((((.....)))))))))(((((.................))).)))))))....)))))))))... (-23.33 = -24.85 + 1.52)

| Location | 14,813,100 – 14,813,211 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 97.01 |
| Mean single sequence MFE | -27.97 |
| Consensus MFE | -25.90 |
| Energy contribution | -26.40 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.857076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14813100 111 + 22224390 ACUGGGGAAAAGUUGGCUGCUUUUCCCGGCCACUUUGUUGCUGGUU-UGGACCGAGUUGAAAGUUUACGCAAUCAACUUUUUCGCCAUUCUCUUUUUUUUUUUAUUUAUCAG ..(((((((((((((..(((.....(((((.((...)).)))))((-(.(((...))).)))......)))..)))))))))).)))......................... ( -28.10) >DroSec_CAF1 7048 111 + 1 ACUGGGGAAAAGUUGGCUGCUUUUCCCGGCCACUUUGUUGCUGGUUUUGGACCGAGUUGAAAGUUUACGCAAUCAACUUUUUCGCCAUUCUCUUUUU-UUUUUAUUUAUCAG ..(((((((((((((..(((.....(((((.((...)).)))))((((.(((...))).)))).....)))..)))))))))).)))..........-.............. ( -28.50) >DroSim_CAF1 6275 112 + 1 ACUGGGGAAAAGUUGGCUGCUUUUCCCGGCCACUUUGUUGCUGGUUUUGGACCGAGUUGAAAGUUUACGCAAUCAACUUUUUCGCCAUUCUCUUUUUUUUUUUAUUUAUCAG ..(((((((((((((..(((.....(((((.((...)).)))))((((.(((...))).)))).....)))..)))))))))).)))......................... ( -28.50) >DroEre_CAF1 12298 110 + 1 ACUGGGGAAAAGUUGGCUGCUUUUCCCGGCCACUUUGUUGCUGCUU-UGGUCCGAGUUGAAAGUUUACGCAAUCAACUUUUUCGCCAUUCCCUUCUU-UUUUUAUUUAUCAG ..(((((((((((((..((((((..(((((((....((....))..-))).))).)..))).(....))))..)))))))))).)))..........-.............. ( -26.80) >consensus ACUGGGGAAAAGUUGGCUGCUUUUCCCGGCCACUUUGUUGCUGGUU_UGGACCGAGUUGAAAGUUUACGCAAUCAACUUUUUCGCCAUUCUCUUUUU_UUUUUAUUUAUCAG .((((((((((((.....)))))))))(((............(((.....)))(((((((..((....))..)))))))....))).......................))) (-25.90 = -26.40 + 0.50)



| Location | 14,813,100 – 14,813,211 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 97.01 |
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -23.81 |
| Energy contribution | -23.81 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14813100 111 - 22224390 CUGAUAAAUAAAAAAAAAAAGAGAAUGGCGAAAAAGUUGAUUGCGUAAACUUUCAACUCGGUCCA-AACCAGCAACAAAGUGGCCGGGAAAAGCAGCCAACUUUUCCCCAGU (((.......................(((.....((((((..(......)..)))))).(((...-.)))............)))((((((((.......))))))))))). ( -24.50) >DroSec_CAF1 7048 111 - 1 CUGAUAAAUAAAAA-AAAAAGAGAAUGGCGAAAAAGUUGAUUGCGUAAACUUUCAACUCGGUCCAAAACCAGCAACAAAGUGGCCGGGAAAAGCAGCCAACUUUUCCCCAGU (((...........-...........(((.....((((((..(......)..)))))).(((.....)))............)))((((((((.......))))))))))). ( -24.50) >DroSim_CAF1 6275 112 - 1 CUGAUAAAUAAAAAAAAAAAGAGAAUGGCGAAAAAGUUGAUUGCGUAAACUUUCAACUCGGUCCAAAACCAGCAACAAAGUGGCCGGGAAAAGCAGCCAACUUUUCCCCAGU (((.......................(((.....((((((..(......)..)))))).(((.....)))............)))((((((((.......))))))))))). ( -24.50) >DroEre_CAF1 12298 110 - 1 CUGAUAAAUAAAAA-AAGAAGGGAAUGGCGAAAAAGUUGAUUGCGUAAACUUUCAACUCGGACCA-AAGCAGCAACAAAGUGGCCGGGAAAAGCAGCCAACUUUUCCCCAGU (((...........-...........(((.(....((((.((((.......(((.....)))...-..))))))))....).)))((((((((.......))))))))))). ( -24.60) >consensus CUGAUAAAUAAAAA_AAAAAGAGAAUGGCGAAAAAGUUGAUUGCGUAAACUUUCAACUCGGUCCA_AACCAGCAACAAAGUGGCCGGGAAAAGCAGCCAACUUUUCCCCAGU .........................(((.(.((((((((..((((........)..(((((.(((...............))))))))....)))..)))))))).)))).. (-23.81 = -23.81 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:52 2006