
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,811,779 – 14,812,032 |
| Length | 253 |
| Max. P | 0.894205 |

| Location | 14,811,779 – 14,811,896 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.25 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -20.16 |
| Energy contribution | -19.84 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889717 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14811779 117 - 22224390 GGCCACCCAUGGGCCACCCAUACCCGUGAAUGGCCCCAUCCCUCCAACGACCAUU--GUGGUCAUUCUUAAAUGGCCUUUUACUUAAUGCUAAGUGUCGUUUGACUGCUGUGCUAUCC-U (((((..((((((.........))))))..))))).......((.((((((....--..(((((((....)))))))....(((((....))))))))))).))..............-. ( -34.20) >DroSec_CAF1 5730 100 - 1 GGCCACCCAUGGGCC-CCCAU-----------------UCCCGCCAGCGACCAUU--GUGGUCAUUCUUAAAUGGCCUUUUACUUAAAGCUAAGUGUCGUUUGACUGCUGUGCUGCCCCA (((.....((((...-.))))-----------------....((((((((((...--..))))....(((((((((.....(((((....))))))))))))))..)))).)).)))... ( -30.50) >DroSim_CAF1 4964 100 - 1 GGCCACCCAUGGGCC-CCCAU-----------------UCCCGCCAGCGACCAUU--GUGGUCAUUCUUAAAUGGCCUUUUACUUAAUGCUAAGUGUCGUUUGACUGCUGUGCUGCCCCA (((.....((((...-.))))-----------------....((((((((((...--..))))....(((((((((.....(((((....))))))))))))))..)))).)).)))... ( -30.50) >DroEre_CAF1 10973 113 - 1 GGCCACCCAGGGGCC-CCCCUCCCCGUGAAUGCCCC---UCCUCCGACGACCAUU--GUGGUCAUUCUUAAAUGGCCUUUUACUUAAUGCUAAGUGUCGUUUGACUGCUGUGCUGCCCC- ((((((..((((((.-...............)))))---)..((.((((((....--..(((((((....)))))))....(((((....))))))))))).)).....)))..)))..- ( -31.39) >DroAna_CAF1 33142 101 - 1 GGUGGCCGA--GACC-CCUCUCUGCG------------UCUC-UCUGCCACCAUUCUGUGACCAUUCUUAAAUGGCCUUUUACUUAAUGCUAAGUGUCGUUUGACGGCUGCC--UCCUC- ((((((.((--((..-((.....).)------------..))-)).)))))).....((..((....(((((((((.....(((((....)))))))))))))).))..)).--.....- ( -27.90) >consensus GGCCACCCAUGGGCC_CCCAU___CG____________UCCCGCCAGCGACCAUU__GUGGUCAUUCUUAAAUGGCCUUUUACUUAAUGCUAAGUGUCGUUUGACUGCUGUGCUGCCCC_ ((((((..((((.....))))......................(.((((((........(((((((....)))))))....(((((....))))))))))).)......)))..)))... (-20.16 = -19.84 + -0.32)



| Location | 14,811,818 – 14,811,934 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.36 |
| Mean single sequence MFE | -38.34 |
| Consensus MFE | -20.59 |
| Energy contribution | -20.35 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14811818 116 + 22224390 AAAGGCCAUUUAAGAAUGACCAC--AAUGGUCGUUGGAGGGAUGGGGCCAUUCACGGGUAUGGGUGGCCCAUGGGUGGCCAAGUCC-AAGUUUCGCCUC-CAUCCAGUCAAAUUGGAUUA ..((((........(((((((..--...)))))))....((((..(((((((((.((((.......)))).)))))))))..))))-.......)))).-.(((((((...))))))).. ( -47.30) >DroSec_CAF1 5770 98 + 1 AAAGGCCAUUUAAGAAUGACCAC--AAUGGUCGCUGGCGGGA-----------------AUGGG-GGCCCAUGGGUGGCCAAGUCC-AAGUUUCGCCUC-CGUCCAGUCAAAUUGGAUUA ...(((((((.............--)))))))(..((((..(-----------------.((((-(((((....).))))...)))-)..)..))))..-)(((((((...))))))).. ( -31.22) >DroSim_CAF1 5004 98 + 1 AAAGGCCAUUUAAGAAUGACCAC--AAUGGUCGCUGGCGGGA-----------------AUGGG-GGCCCAUGGGUGGCCAAGUCC-AAGUUUCGCCUC-CGUCCAGUCAAAUUGGAUUA ...(((((((.............--)))))))(..((((..(-----------------.((((-(((((....).))))...)))-)..)..))))..-)(((((((...))))))).. ( -31.22) >DroEre_CAF1 11012 113 + 1 AAAGGCCAUUUAAGAAUGACCAC--AAUGGUCGUCGGAGGA---GGGGCAUUCACGGGGAGGGG-GGCCCCUGGGUGGCCAAGUCCCAUAUUUCGCCUC-CGUCCAGUCAAAUUGGAUUA .....(((.....((.(((((..--...)))))))(((.(.---(((((.......((((....-(((((....).))))...)))).......)))))-).)))........))).... ( -40.04) >DroAna_CAF1 33179 103 + 1 AAAGGCCAUUUAAGAAUGGUCACAGAAUGGUGGCAGA-GAGA------------CGCAGAGAGG-GGUC--UCGGCCACCAAGUCC-AAGUUUCGCCUUUUGGCCACUCAAAUUGGAUUA ...(((((((....)))))))......(((((((...-((((------------(.(......)-.)))--)).)))))))(((((-((.....(((....)))........))))))). ( -41.92) >consensus AAAGGCCAUUUAAGAAUGACCAC__AAUGGUCGCUGGAGGGA____________CG___AUGGG_GGCCCAUGGGUGGCCAAGUCC_AAGUUUCGCCUC_CGUCCAGUCAAAUUGGAUUA ..((((((((....)))).........((((((((.(.(((..........................))).).)))))))).............))))...(((((((...))))))).. (-20.59 = -20.35 + -0.24)

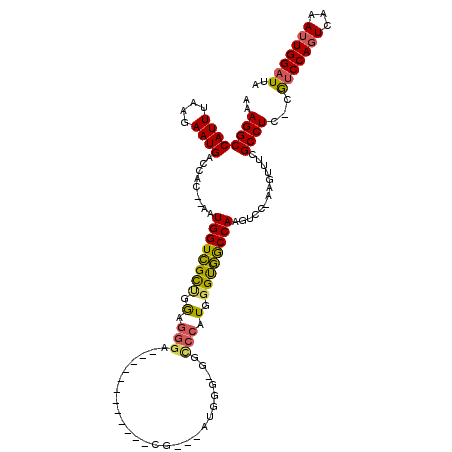
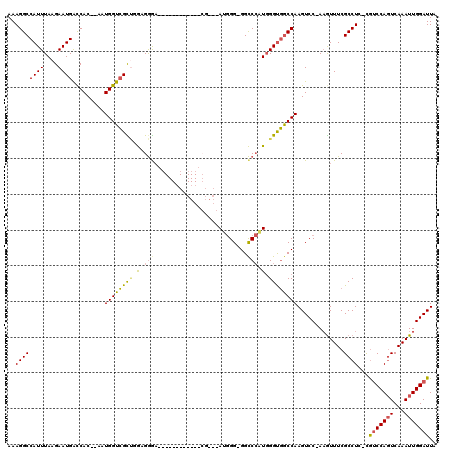
| Location | 14,811,818 – 14,811,934 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.36 |
| Mean single sequence MFE | -33.32 |
| Consensus MFE | -19.06 |
| Energy contribution | -19.42 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14811818 116 - 22224390 UAAUCCAAUUUGACUGGAUG-GAGGCGAAACUU-GGACUUGGCCACCCAUGGGCCACCCAUACCCGUGAAUGGCCCCAUCCCUCCAACGACCAUU--GUGGUCAUUCUUAAAUGGCCUUU ..(((((.......)))))(-((((.((....(-((....(((((..((((((.........))))))..)))))))))))))))..........--..(((((((....)))))))... ( -39.80) >DroSec_CAF1 5770 98 - 1 UAAUCCAAUUUGACUGGACG-GAGGCGAAACUU-GGACUUGGCCACCCAUGGGCC-CCCAU-----------------UCCCGCCAGCGACCAUU--GUGGUCAUUCUUAAAUGGCCUUU .....(((((((.((((.((-(.(((.((....-....)).)))....((((...-.))))-----------------..))))))))))..)))--).(((((((....)))))))... ( -29.40) >DroSim_CAF1 5004 98 - 1 UAAUCCAAUUUGACUGGACG-GAGGCGAAACUU-GGACUUGGCCACCCAUGGGCC-CCCAU-----------------UCCCGCCAGCGACCAUU--GUGGUCAUUCUUAAAUGGCCUUU .....(((((((.((((.((-(.(((.((....-....)).)))....((((...-.))))-----------------..))))))))))..)))--).(((((((....)))))))... ( -29.40) >DroEre_CAF1 11012 113 - 1 UAAUCCAAUUUGACUGGACG-GAGGCGAAAUAUGGGACUUGGCCACCCAGGGGCC-CCCCUCCCCGUGAAUGCCCC---UCCUCCGACGACCAUU--GUGGUCAUUCUUAAAUGGCCUUU ....(((.(((((.((((.(-((((.(...((((((....((((.(....)))))-......))))))....).))---)))))))..((((...--..))))....))))))))..... ( -36.70) >DroAna_CAF1 33179 103 - 1 UAAUCCAAUUUGAGUGGCCAAAAGGCGAAACUU-GGACUUGGUGGCCGA--GACC-CCUCUCUGCG------------UCUC-UCUGCCACCAUUCUGUGACCAUUCUUAAAUGGCCUUU ....(((.((.((((((.((.(((......)))-(((..(((((((.((--((..-((.....).)------------..))-)).))))))).))).)).))))))..)).)))..... ( -31.30) >consensus UAAUCCAAUUUGACUGGACG_GAGGCGAAACUU_GGACUUGGCCACCCAUGGGCC_CCCAU___CG____________UCCCGCCAGCGACCAUU__GUGGUCAUUCUUAAAUGGCCUUU ...((((.......))))...(((((........((....((((.......))))..)).............................((((((...))))))...........))))). (-19.06 = -19.42 + 0.36)

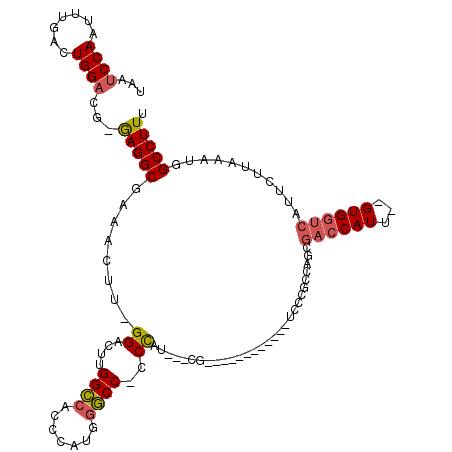
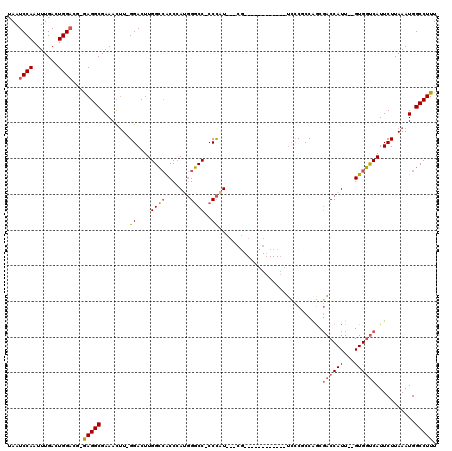
| Location | 14,811,934 – 14,812,032 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.07 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -12.58 |
| Energy contribution | -13.90 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14811934 98 - 22224390 ---UCUGUCACUU--UCAGUUGGCCCCUGUUU--------------UGUGCCCCCCGCCCA-GGCACACCUG-CCAGGCACUUCACACUGACAUGGGC-GAUGGGAAUGGGGCAACAACU ---..........--..((((((((((.....--------------....(((..((((((-((((....))-)).......((.....))..)))))-)..)))...)))))..))))) ( -33.80) >DroSec_CAF1 5868 98 - 1 ---UCUGCCACUU--UCAGUAGGCCCCUGUUU--------------UGUGCCCCCCGCCCA-GGCACACCUG-CCAGGCACUUCACACUGACAUGGGC-GAUGGGGAUGGGGCAACAACU ---.((((.....--...))))(((((.....--------------....((((.((((((-((((....))-)).......((.....))..)))))-)..))))..)))))....... ( -35.90) >DroSim_CAF1 5102 98 - 1 ---UCUGCCACUU--UCGGUAGGCCCCUGUUU--------------CGUGCCCCCCGGCCA-GGCACACCUG-CCAGGCACUUCACACUGACAUGGGC-GAUGGGGAUGGGGCAACAACU ---.(((((....--..)))))(((((..(..--------------(((((((....(((.-((((....))-)).)))...((.....))...))))-.)))..)..)))))....... ( -39.90) >DroEre_CAF1 11125 112 - 1 ---UCUGCCACUU--UCAGUGGGUCCCCGUUUACAGACCCCCACUUUCAGCCCCCCACACA-GGCACACCUG-CCAGGCACUUCACAGUGACAUGGAA-GAUGGGGAUGGGACAACAACU ---....((((..--...))))(((((.(((....)))(((((((((((............-((((....))-)).(.((((....)))).).)))))-).)))))..)))))....... ( -37.30) >DroAna_CAF1 33282 91 - 1 CAUUCUCUCAUUUCGUCAGUUGGCCAAGCGAG--------------UG--------GGCCAGAGAACACCUGGCCAGAAAC-------CGACAACCAAAAGAGGAGAAGGGGCAACAACU .....((((.((((.((..((((.....((.(--------------(.--------((((((.......))))))....))-------))....))))..)).)))).))))........ ( -27.60) >consensus ___UCUGCCACUU__UCAGUAGGCCCCUGUUU______________UGUGCCCCCCGCCCA_GGCACACCUG_CCAGGCACUUCACACUGACAUGGGC_GAUGGGGAUGGGGCAACAACU .................(((..(((((....................(((((..........))))).(((....)))..............................)))))....))) (-12.58 = -13.90 + 1.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:48 2006