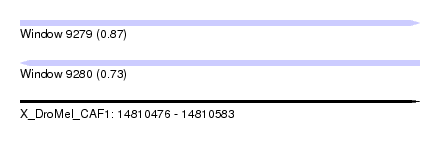
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,810,476 – 14,810,583 |
| Length | 107 |
| Max. P | 0.873738 |

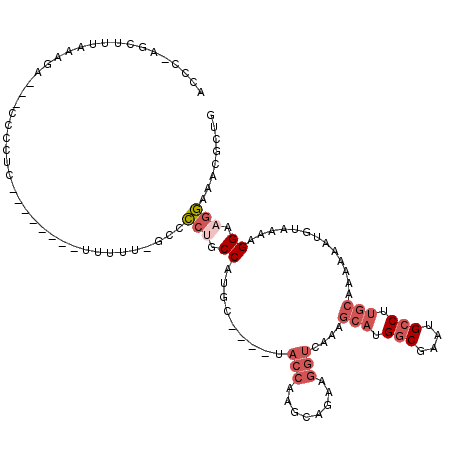
| Location | 14,810,476 – 14,810,583 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.15 |
| Mean single sequence MFE | -31.94 |
| Consensus MFE | -8.68 |
| Energy contribution | -10.36 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.27 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14810476 107 + 22224390 ACCC-AGCUUUAAAGA---CUCCAC--------GUUUU-GCCCCUGCCAUGCUGCCUACCAAGCAGAAGGUCAAAGCAUGGCGAAUGCCUUGCAAAAAAUGUAAAAGGAAGGAAACGCUG ...(-(((........---.(((..--------.....-((...(((((((((....(((........)))...)))))))))...)).(((((.....)))))..))).(....))))) ( -30.30) >DroSec_CAF1 4531 107 + 1 GCCC-AGCUUUAAAGAGCACCCCUC--------UUUUUGACCCCUGCCAUGC----UACCAAGCAGAAGGUCAAAGCAUGGCGAAUGCCUUGCAAAAAAUGUAAAAGGAAGGAAACGCUG ...(-(((...((((((.....)))--------)))......((((((((((----((((........)))...)))))))).....(((((((.....))))..))).)))....)))) ( -30.40) >DroSim_CAF1 3736 115 + 1 GCCC-AGCUUUAAAGAGCACCCCCCUCCCUCCUUUUUUGACCCCUGCCACGC----UACCAAGCAGAAGGUCAAAGCAUGGCGAAUGCCUUGCAAAAAAUGUAAAAGGAAGGAAACGCUG ((..-.((((....)))).......(((.(((((((((((((.((((.....----......))))..)))))))(((.(((....))).)))..........)))))).)))...)).. ( -35.20) >DroEre_CAF1 9718 103 + 1 ACCC-AGCUUUAAAGA---CCCCUC--------CUUUU-GCUCCCGCCACGC----UACCGAGCAGAAGGUCAAAGCAUGGCGAGUGCCUUGCAAAAAAUGUAAAAGGAAGGAAACGCUG ...(-(((........---....((--------(((((-((...(((((.((----((((........)))...))).)))))..(((...)))......))))))))).(....))))) ( -32.70) >DroAna_CAF1 32088 90 + 1 ACCCCAGAGUCGGAGG------------------AGUC-GCUGCCUCCAUUUUGC--------GAGAAGGUCAAAG--UGGCAAGCGGCCUGGGGGAUGGGGGGCAGGAACCAAACACA- .((((((..((....)------------------)(((-(((....(((((((((--------......).)))))--)))..))))))))))))..((..((.......))...))..- ( -31.10) >consensus ACCC_AGCUUUAAAGA___CCCCUC________UUUUU_GCCCCUGCCAUGC____UACCAAGCAGAAGGUCAAAGCAUGGCGAAUGCCUUGCAAAAAAUGUAAAAGGAAGGAAACGCUG ..........................................(((.((.........(((........)))....(((.(((....))).))).............)).)))........ ( -8.68 = -10.36 + 1.68)


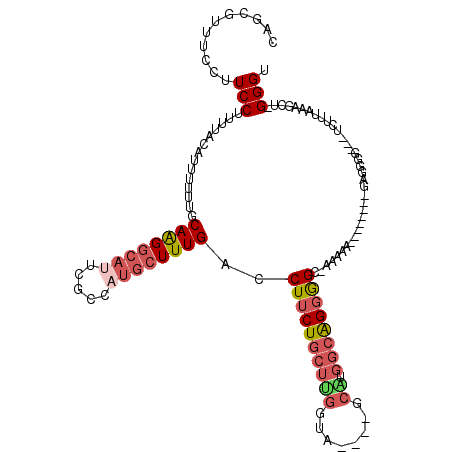
| Location | 14,810,476 – 14,810,583 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.15 |
| Mean single sequence MFE | -35.87 |
| Consensus MFE | -13.06 |
| Energy contribution | -14.34 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.727483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14810476 107 - 22224390 CAGCGUUUCCUUCCUUUUACAUUUUUUGCAAGGCAUUCGCCAUGCUUUGACCUUCUGCUUGGUAGGCAGCAUGGCAGGGGC-AAAAC--------GUGGAG---UCUUUAAAGCU-GGGU ((((.........(((...(((.((((((..(((....)))...(((((.((..(((((.....)))))...)))))))))-)))).--------))))))---........)))-)... ( -30.53) >DroSec_CAF1 4531 107 - 1 CAGCGUUUCCUUCCUUUUACAUUUUUUGCAAGGCAUUCGCCAUGCUUUGACCUUCUGCUUGGUA----GCAUGGCAGGGGUCAAAAA--------GAGGGGUGCUCUUUAAAGCU-GGGC ((((((.((((((..............(((.(((....))).)))((((((((.(((((((...----.)).)))))))))))))..--------)))))).))........)))-)... ( -35.90) >DroSim_CAF1 3736 115 - 1 CAGCGUUUCCUUCCUUUUACAUUUUUUGCAAGGCAUUCGCCAUGCUUUGACCUUCUGCUUGGUA----GCGUGGCAGGGGUCAAAAAAGGAGGGAGGGGGGUGCUCUUUAAAGCU-GGGC ((((...(((((((((((.........(((.(((....))).))).(((((((.(((((.....----....)))))))))))))))))))))))((((....)))).....)))-)... ( -46.40) >DroEre_CAF1 9718 103 - 1 CAGCGUUUCCUUCCUUUUACAUUUUUUGCAAGGCACUCGCCAUGCUUUGACCUUCUGCUCGGUA----GCGUGGCGGGAGC-AAAAG--------GAGGGG---UCUUUAAAGCU-GGGU ((((...(((((((((((.((.....))....((.(((((((((((((((.(....).)))).)----)))))))))).))-)))))--------))))))---........)))-)... ( -42.20) >DroAna_CAF1 32088 90 - 1 -UGUGUUUGGUUCCUGCCCCCCAUCCCCCAGGCCGCUUGCCA--CUUUGACCUUCUC--------GCAAAAUGGAGGCAGC-GACU------------------CCUCCGACUCUGGGGU -.(((...(((....)))...))).((((((((.((((.(((--.((((.(......--------))))).))))))).))-((..------------------..)).....)))))). ( -24.30) >consensus CAGCGUUUCCUUCCUUUUACAUUUUUUGCAAGGCAUUCGCCAUGCUUUGACCUUCUGCUUGGUA____GCAUGGCAGGGGC_AAAAA________GAGGGG___UCUUUAAAGCU_GGGU ...........(((..............((((((((.....))))))))..((((((((((........)).))))))))....................................))). (-13.06 = -14.34 + 1.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:43 2006