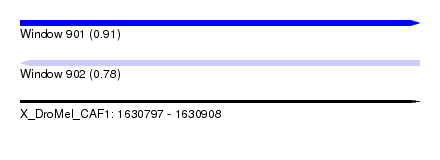
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,630,797 – 1,630,908 |
| Length | 111 |
| Max. P | 0.905328 |

| Location | 1,630,797 – 1,630,908 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.59 |
| Mean single sequence MFE | -31.64 |
| Consensus MFE | -25.70 |
| Energy contribution | -25.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1630797 111 + 22224390 AAGAAGGGAAAGGCUUCACUGCUGCAUUGCAGUGAAUUGCGUUCAUCUGCCCC--------UCGCGAACUU-CCCCUGUUCACUUUGCAUUUCACUUUCUUGAGCGAAACAAACAAGCCA (((((((((((.((((((((((......))))))))..(((............--------.)))((((..-.....)))).....)).)))).)))))))(.((...........))). ( -29.42) >DroSec_CAF1 25919 110 + 1 AAGAAGGGAAAGGCUUCACUGCUGCAUUGCAGUGAAUUGCGUUCAUCUGCCCC--------UCGCGCCCUU-CCCC-AUUCACUUUGCAUUUCACUUUCUUGAGCGAAACAAACAAGCCA ..((((((((.(((((((((((......))))))))..(((............--------.)))))).))-))).-.)))..((((..((((.(((....))).))))))))....... ( -32.42) >DroSim_CAF1 42444 110 + 1 AAGAAGGGAAAGGCUUCACUGCUGCAUUGCAGUGAAUUGCGUUCAUCUGCCCC--------UCGCGCUCCU-CCCC-AUUCACUUUGCAUUUCACUUUCUUGAGCGAAACAAACAAGCCA ..(((((((..(((((((((((......))))))))..(((............--------.))))))..)-))).-.)))..((((..((((.(((....))).))))))))....... ( -29.22) >DroYak_CAF1 30238 119 + 1 AAGAAGGGAAAGGCUUCACUGCUGCAUUGCAGUGAAUUGCGUUCAUCUGCCCCUUCGCCCCUCGCGCCACUACCCC-GUUCAAUUUGCAUUUCCCUUUCUUGGGCGAAACAAACAAGCCA ((((((((((((((((((((((......))))))))..(((.......((......))....))))))........-((.......)).)))))).))))).(((...........))). ( -35.50) >consensus AAGAAGGGAAAGGCUUCACUGCUGCAUUGCAGUGAAUUGCGUUCAUCUGCCCC________UCGCGCCCCU_CCCC_AUUCACUUUGCAUUUCACUUUCUUGAGCGAAACAAACAAGCCA (((((((((((.((((((((((......))))))))..(((.....................))).....................)).)))).)))))))(.((...........))). (-25.70 = -25.70 + -0.00)

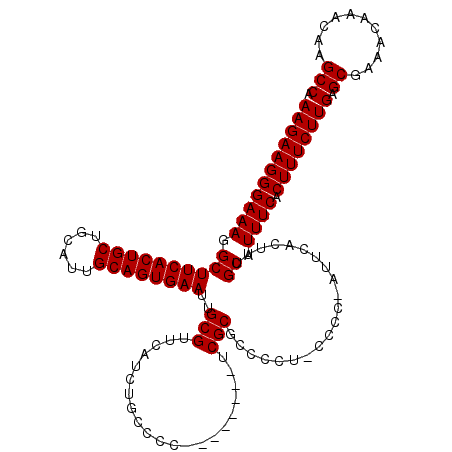
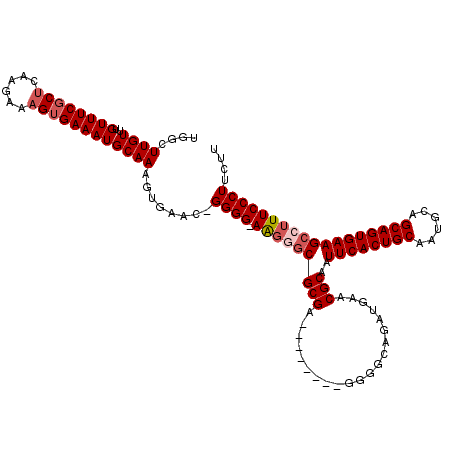
| Location | 1,630,797 – 1,630,908 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.59 |
| Mean single sequence MFE | -39.21 |
| Consensus MFE | -29.38 |
| Energy contribution | -30.62 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1630797 111 - 22224390 UGGCUUGUUUGUUUCGCUCAAGAAAGUGAAAUGCAAAGUGAACAGGGG-AAGUUCGCGA--------GGGGCAGAUGAACGCAAUUCACUGCAAUGCAGCAGUGAAGCCUUUCCCUUCUU .((.((((..((((((((......)))))))))))).((((((.....-..))))))((--------(((((...((....)).((((((((......)))))))))))))))))..... ( -36.60) >DroSec_CAF1 25919 110 - 1 UGGCUUGUUUGUUUCGCUCAAGAAAGUGAAAUGCAAAGUGAAU-GGGG-AAGGGCGCGA--------GGGGCAGAUGAACGCAAUUCACUGCAAUGCAGCAGUGAAGCCUUUCCCUUCUU (.((((...(((((((((......)))))))))..)))).)..-((((-(((((((((.--------............)))..((((((((......)))))))))))))))))).... ( -42.22) >DroSim_CAF1 42444 110 - 1 UGGCUUGUUUGUUUCGCUCAAGAAAGUGAAAUGCAAAGUGAAU-GGGG-AGGAGCGCGA--------GGGGCAGAUGAACGCAAUUCACUGCAAUGCAGCAGUGAAGCCUUUCCCUUCUU ..((((...(((((((((......)))))))))..))))(((.-((((-(((.(((((.--------............)))..((((((((......)))))))))))))))))))).. ( -38.72) >DroYak_CAF1 30238 119 - 1 UGGCUUGUUUGUUUCGCCCAAGAAAGGGAAAUGCAAAUUGAAC-GGGGUAGUGGCGCGAGGGGCGAAGGGGCAGAUGAACGCAAUUCACUGCAAUGCAGCAGUGAAGCCUUUCCCUUCUU ..(((.((((((....(((......)))....))))))...((-......)))))..((((((.(((((.((........))..((((((((......)))))))).))))))))))).. ( -39.30) >consensus UGGCUUGUUUGUUUCGCUCAAGAAAGUGAAAUGCAAAGUGAAC_GGGG_AAGGGCGCGA________GGGGCAGAUGAACGCAAUUCACUGCAAUGCAGCAGUGAAGCCUUUCCCUUCUU ....((((..((((((((......))))))))))))........((((.(((((((((.....................)))..((((((((......)))))))))))))))))).... (-29.38 = -30.62 + 1.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:04 2006