
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,792,450 – 14,792,609 |
| Length | 159 |
| Max. P | 0.781595 |

| Location | 14,792,450 – 14,792,547 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 74.23 |
| Mean single sequence MFE | -26.91 |
| Consensus MFE | -19.61 |
| Energy contribution | -21.18 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14792450 97 + 22224390 CGUCGCUUCCGUUUCCGGAUGCUGCUGCACUUCCCCUGCGCACC------CUGCUCUUCAAC--------UAACUGCAUUAUCGCUUAUCUACACUGAGCGGAAAUGCGGU ....((.((((....)))).))(((.(((.......))))))..------............--------..((((((((.(((((((.......))))))).)))))))) ( -28.70) >DroSec_CAF1 27263 105 + 1 CGUCGCUUCCGUUUCCGGAUGGUGCUGCACUUCCCCUGCGCACC------CUGCUCCCCUACUCUUCUCAUAACUGCAUUAUCGCUUAUCAACACUGAGCGGAAAUACGGU ....((.((((....)))).(((((.(((.......))))))))------..))..................((((.(((.(((((((.......))))))).))).)))) ( -26.30) >DroEre_CAF1 22525 111 + 1 CGUGGCUUCCGUUUCCGGAUGGUGCUGCAUUACAGCCACCCACCUCGCCCCUGGCCCACUGCUAUUCUGCUAACUGCAUUAUCGCUUAUCUACACUGAGCGGAAAGGCGCU .((((..((((....)))).(((((((.....))).))))......(((...))))))).(((.((((((.....)))....((((((.......))))))))).)))... ( -31.90) >DroYak_CAF1 22201 88 + 1 CGUGGCUUCCGUUUCCGGAUGACGUUGCAUUU-----------------------CCCCUGCUCUUUUACUAACUGCAUUAUCGCUUAUCUACACUGAGCGGAAAGGCGCU .(((.(((((((((..((((((((.((((...-----------------------...................))))....)).)))))).....)))))).))).))). ( -20.75) >consensus CGUCGCUUCCGUUUCCGGAUGGUGCUGCACUUCCCCUGCGCACC______CUGCUCCCCUACUCUUCU_CUAACUGCAUUAUCGCUUAUCUACACUGAGCGGAAAGGCGCU .(((((.((((....))))....)))))............................................((((((((.(((((((.......))))))).)))))))) (-19.61 = -21.18 + 1.56)



| Location | 14,792,481 – 14,792,579 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 68.24 |
| Mean single sequence MFE | -22.67 |
| Consensus MFE | -13.62 |
| Energy contribution | -13.73 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14792481 98 + 22224390 UCCCCUGCGCACC------CUGCUCUUCAAC--------UAACUGCAUUAUCGCUUAUCUACACUGAGCGGAAAUGCGGUAAUUCAUAAAUGAUUUGCUGCUUC----GAUUG-GCU .............------..(((..((...--------.....(((((.(((((((.......))))))).)))))(((((.(((....))).))))).....----))..)-)). ( -23.70) >DroSec_CAF1 27294 106 + 1 UCCCCUGCGCACC------CUGCUCCCCUACUCUUCUCAUAACUGCAUUAUCGCUUAUCAACACUGAGCGGAAAUACGGUCAUUCAUAAAUGAUUUGUUGUUUC----UUUUG-GCU ........((...------..)).....................((.....((((((.......))))))((((((((((((((....))))).))).))))))----.....-)). ( -15.60) >DroEre_CAF1 22556 110 + 1 ACAGCCACCCACCUCGCCCCUGGCCCACUGCUAUUCUGCUAACUGCAUUAUCGCUUAUCUACACUGAGCGGAAAGGCGCUCAUUCAUCAU------GCAGUUUGCAGUGAUUUA-CU ...((((.............)))).((((((....((((.....((......))..........((((((......))))))........------))))...)))))).....-.. ( -28.72) >consensus UCCCCUGCGCACC______CUGCUCCACUACU_UUCU__UAACUGCAUUAUCGCUUAUCUACACUGAGCGGAAAUGCGGUCAUUCAUAAAUGAUUUGCUGUUUC____GAUUG_GCU ......(((((.........)))..................((((((((.(((((((.......))))))).))))))))...................))................ (-13.62 = -13.73 + 0.11)


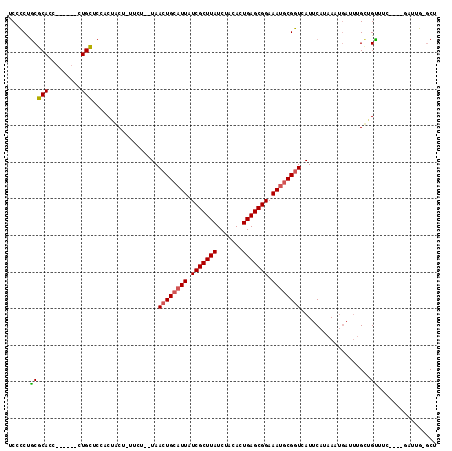
| Location | 14,792,507 – 14,792,609 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 71.08 |
| Mean single sequence MFE | -22.81 |
| Consensus MFE | -11.27 |
| Energy contribution | -12.53 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
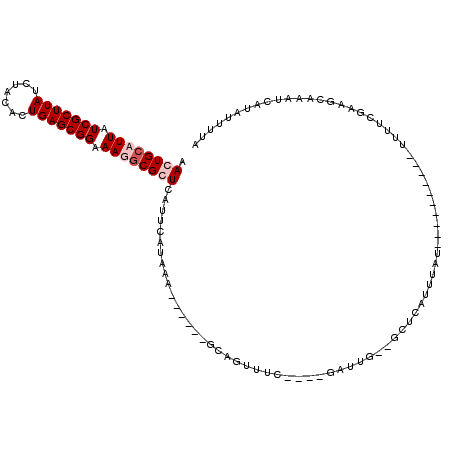
>X_DroMel_CAF1 14792507 102 + 22224390 AACUGCAUUAUCGCUUAUCUACACUGAGCGGAAAUGCGGUAAUUCAUAAAUGAUUUGCUGCUUC----GAUUG--GCUCAUUUAU----------UUUUCGAAGCAAAUCAUAUUUCA .((((((((.(((((((.......))))))).)))))))).........((((((((((...((----((..(--(........)----------)..))))))))))))))...... ( -31.90) >DroSec_CAF1 27328 102 + 1 AACUGCAUUAUCGCUUAUCAACACUGAGCGGAAAUACGGUCAUUCAUAAAUGAUUUGUUGUUUC----UUUUG--GCUCAUUUAU----------UUUUCGAAGCAAAUCAUAUUUGA .((((.(((.(((((((.......))))))).))).))))...(((...((((((((((.....----.....--..........----------.......))))))))))...))) ( -19.93) >DroEre_CAF1 22596 108 + 1 AACUGCAUUAUCGCUUAUCUACACUGAGCGGAAAGGCGCUCAUUCAUCAU------GCAGUUUGCAGUGAUUUA--CUCAUUUUUCUGGCCCCGCUAUUAGCACUCAAUUA--UUUUU ((((((((....(........)..((((((......))))))......))------))))))((((((((....--.)))))....((((...))))...)))........--..... ( -20.00) >DroYak_CAF1 22249 102 + 1 AACUGCAUUAUCGCUUAUCUACACUGAGCGGAAAGGCGCUUAUUCAUCAU------GCAGUUUUAUGUGAUUUAUAAUCAUUUAG----------UUUUCAAAGUAAACUAGGUUUUA ((((((((.(((((((.((..(.....)..)).))))).......)).))------))))))....(((((.....)))))((((----------(((.......)))))))...... ( -19.41) >consensus AACUGCAUUAUCGCUUAUCUACACUGAGCGGAAAGGCGCUCAUUCAUAAA______GCAGUUUC____GAUUG__GCUCAUUUAU__________UUUUCGAAGCAAAUCAUAUUUUA .((((((((.(((((((.......))))))).)))))))).............................................................................. (-11.27 = -12.53 + 1.25)

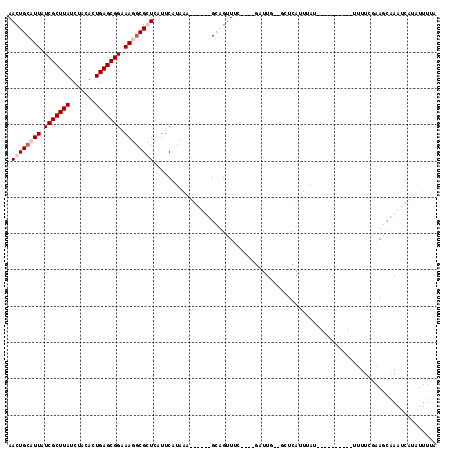
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:35 2006