
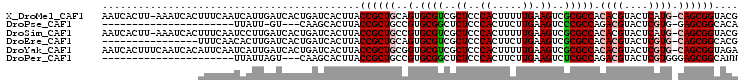
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,784,211 – 14,784,315 |
| Length | 104 |
| Max. P | 0.973641 |

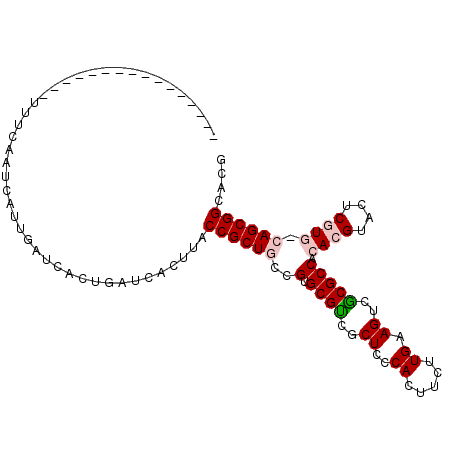
| Location | 14,784,211 – 14,784,315 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 74.60 |
| Mean single sequence MFE | -26.92 |
| Consensus MFE | -14.92 |
| Energy contribution | -15.45 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14784211 104 + 22224390 AAUCACUU-AAAUCACUUUCAAUCAUUGAUCACUGAUCACUUACCGCUGCAGUGCGUCGCUCCCACUUUUUGAAGUCGCGCCACACGUACUCAUG-CAGCGGUACG ........-.................(((((...)))))..(((((((((((((((((((....((((....)))).)))....))))))...))-)))))))).. ( -29.40) >DroPse_CAF1 19116 79 + 1 ----------------------UUAUU-GU---CAAGCACUUACCGCUGCCGUGCGGCUCUCCCACUUCUUGAAGUCCCGCCAGACGUACUCGUG-GAGCGGCACA ----------------------.....-..---..(((.......)))...((((.((((....((((....))))..(((.((.....)).)))-)))).)))). ( -19.90) >DroSim_CAF1 19562 104 + 1 AAUCACUU-AAAUCACUUUCAAUCCUUGAUCACUGAUCACUUACCGCUGCCGUGCGUCGCUCCCACUUUUUGAAGUCGCGCCACACGUACUCAUG-CAGCGGUACG ........-.................(((((...)))))..(((((((((.(((((((((....((((....)))).)))....))))))....)-)))))))).. ( -27.70) >DroEre_CAF1 13872 89 + 1 ----------------UUUCAACACUUGAUCACUGAUCACUUACCGCUGCAGUGCGUCGCUCCCACUUCUUGAAGUCGCGCCACACGUACUCGUG-CAGCGGCACG ----------------..........(((((...)))))....(((((((((((((((((....((((....)))).)))....))))))...))-)))))).... ( -27.30) >DroYak_CAF1 13827 105 + 1 AAUCACUUUCAAUCACAUUCAAUCAUUGAUCACUGAUCACUUACCGCUGCGGUGCGUCGCUCCCACUUUUUGAAGUCGCGCCACACGUACUCGUG-CAGCGGUAGA ..........................(((((...))))).((((((((((((((((.(..((.........)).).))))))..(((....))))-))))))))). ( -32.90) >DroPer_CAF1 19666 81 + 1 ----------------------UUAUUAGU---CAAGCACUUACCGCUGCCGUGCGGCUCUCCCACUUCUUGAAGUCUCGCCAGACGUACUCGUGGGAGCGGCANN ----------------------........---...((.....((((......))))(.(((((((........((((....))))......))))))).)))... ( -24.34) >consensus ________________UUUCAAUCAUUGAUCACUGAUCACUUACCGCUGCCGUGCGUCGCUCCCACUUCUUGAAGUCGCGCCACACGUACUCGUG_CAGCGGCACG ...........................................((((((..(.((((..((..((.....)).))..))))).((((....)))).)))))).... (-14.92 = -15.45 + 0.53)
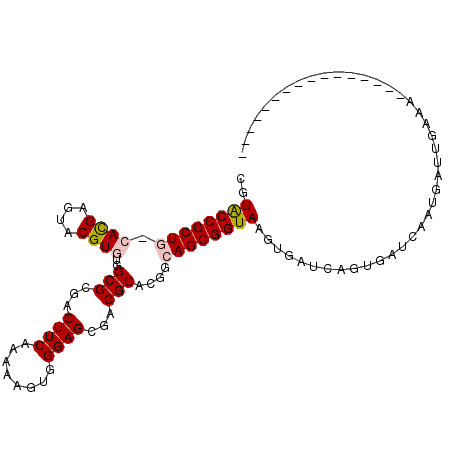
| Location | 14,784,211 – 14,784,315 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 74.60 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -19.06 |
| Energy contribution | -19.25 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14784211 104 - 22224390 CGUACCGCUG-CAUGAGUACGUGUGGCGCGACUUCAAAAAGUGGGAGCGACGCACUGCAGCGGUAAGUGAUCAGUGAUCAAUGAUUGAAAGUGAUUU-AAGUGAUU ..((((((((-((.......(((((.(((..((.(.....).))..))).)))))))))))))))...(((((.((((((.(.......).))))).-.).))))) ( -33.81) >DroPse_CAF1 19116 79 - 1 UGUGCCGCUC-CACGAGUACGUCUGGCGGGACUUCAAGAAGUGGGAGAGCCGCACGGCAGCGGUAAGUGCUUG---AC-AAUAA---------------------- ..((((((((-((((....)))...((((..((((........))))..))))..)).)))))))........---..-.....---------------------- ( -24.90) >DroSim_CAF1 19562 104 - 1 CGUACCGCUG-CAUGAGUACGUGUGGCGCGACUUCAAAAAGUGGGAGCGACGCACGGCAGCGGUAAGUGAUCAGUGAUCAAGGAUUGAAAGUGAUUU-AAGUGAUU ..((((((((-(.......((((((.(((..((.(.....).))..))).)))))))))))))))...(((((.(..(((.....))).).))))).-........ ( -34.51) >DroEre_CAF1 13872 89 - 1 CGUGCCGCUG-CACGAGUACGUGUGGCGCGACUUCAAGAAGUGGGAGCGACGCACUGCAGCGGUAAGUGAUCAGUGAUCAAGUGUUGAAA---------------- ..((((((((-((.......(((((.(((..((.(.....).))..))).)))))))))))))))..(((((...)))))..........---------------- ( -32.71) >DroYak_CAF1 13827 105 - 1 UCUACCGCUG-CACGAGUACGUGUGGCGCGACUUCAAAAAGUGGGAGCGACGCACCGCAGCGGUAAGUGAUCAGUGAUCAAUGAUUGAAUGUGAUUGAAAGUGAUU ..((((((((-(..(....)(((((.(((..((.(.....).))..))).))))).)))))))))...(((((.(..(((((.((.....)).))))).).))))) ( -36.80) >DroPer_CAF1 19666 81 - 1 NNUGCCGCUCCCACGAGUACGUCUGGCGAGACUUCAAGAAGUGGGAGAGCCGCACGGCAGCGGUAAGUGCUUG---ACUAAUAA---------------------- ..((((((((((((......((((....))))........))))))..(((....))).))))))........---........---------------------- ( -27.44) >consensus CGUACCGCUG_CACGAGUACGUGUGGCGCGACUUCAAAAAGUGGGAGCGACGCACGGCAGCGGUAAGUGAUCAGUGAUCAAUGAUUGAAA________________ ..((((((((.((((....))))..(((...((((........))))...)))....))))))))......................................... (-19.06 = -19.25 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:31 2006