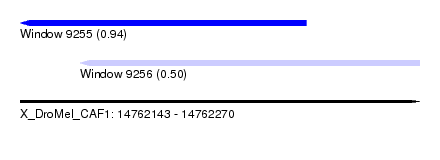
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,762,143 – 14,762,270 |
| Length | 127 |
| Max. P | 0.938388 |

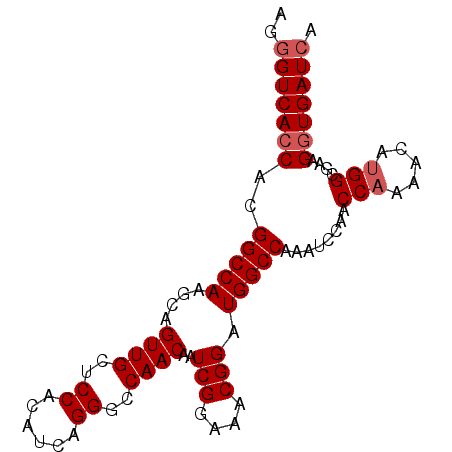
| Location | 14,762,143 – 14,762,234 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 97.07 |
| Mean single sequence MFE | -33.64 |
| Consensus MFE | -29.43 |
| Energy contribution | -30.43 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.94 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14762143 91 - 22224390 AGGGUCACCACGGCCAAGCAGUUGCUCCACAUCAGGGUCAACAAUCGGAAACGGAUGGCCAAAUCCAACCAAAACAUGGCGAAGGUGAUCA ..(((((((..(((((....((((..((......))..))))..(((....))).)))))........(((.....)))....))))))). ( -34.30) >DroSec_CAF1 6841 91 - 1 AGGGUCAGCACGGCCAAGCAGUUGCUCCACAUCAGGGCCAACAAUCGGAAACGGAUGGCCAAAUCCAACCCAAACAUGGCGAAGGUGAUGA .((((......(((((....((((..((......))..))))..(((....))).))))).......))))...(((.((....)).))). ( -32.32) >DroSim_CAF1 6843 91 - 1 AGGGUCACCACGGCCAAGCAGUUGCUCCACAUCAGGGCCAACAAUCGGAAACGGAUGGCCAAAUCCAACCAAAACAUGGCGAAGGUGAUCA ..(((((((..(((((....((((..((......))..))))..(((....))).)))))........(((.....)))....))))))). ( -34.30) >consensus AGGGUCACCACGGCCAAGCAGUUGCUCCACAUCAGGGCCAACAAUCGGAAACGGAUGGCCAAAUCCAACCAAAACAUGGCGAAGGUGAUCA ..(((((((..(((((....((((..((......))..))))..(((....))).)))))........(((.....)))....))))))). (-29.43 = -30.43 + 1.00)


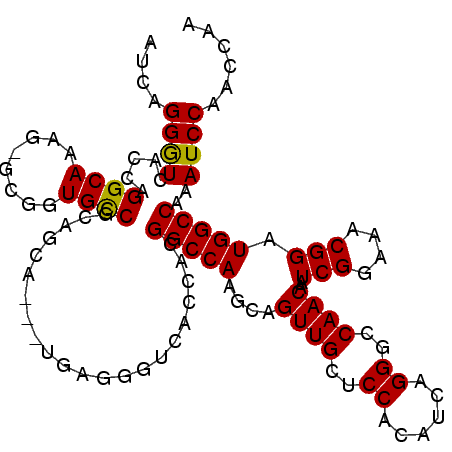
| Location | 14,762,162 – 14,762,270 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 91.57 |
| Mean single sequence MFE | -37.97 |
| Consensus MFE | -30.14 |
| Energy contribution | -29.70 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.08 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14762162 108 - 22224390 AUCAGGAUCACCAGGCAAAG-GAGGUGUCACAGCA---UCAGGGUCACCACGGCCAAGCAGUUGCUCCACAUCAGGGUCAACAAUCGGAAACGGAUGGCCAAAUCCAACCAA ....((....)).((....(-(((((((....)))---)).((....))..(((((....((((..((......))..))))..(((....))).)))))...)))..)).. ( -31.60) >DroSec_CAF1 6860 109 - 1 AUCAGGGUCACCAGGCAAAGGGCGGUGCCGCAGCA---UGAGGGUCAGCACGGCCAAGCAGUUGCUCCACAUCAGGGCCAACAAUCGGAAACGGAUGGCCAAAUCCAACCCA ....((((.....((((........))))(((((.---((..((((.....))))...)))))))..........(((((....(((....))).))))).......)))). ( -37.40) >DroSim_CAF1 6862 111 - 1 AUCAGGGUGACCAGGCAAAG-GCGGUGCCGCAGCCUGGUGAGGGUCACCACGGCCAAGCAGUUGCUCCACAUCAGGGCCAACAAUCGGAAACGGAUGGCCAAAUCCAACCAA ....((((.(((((((....-(((....))).)))))))..((....))..(((((....((((..((......))..))))..(((....))).)))))..))))...... ( -44.90) >consensus AUCAGGGUCACCAGGCAAAG_GCGGUGCCGCAGCA___UGAGGGUCACCACGGCCAAGCAGUUGCUCCACAUCAGGGCCAACAAUCGGAAACGGAUGGCCAAAUCCAACCAA ....((((.....((((........))))......................(((((....((((..((......))..))))..(((....))).)))))..))))...... (-30.14 = -29.70 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:22 2006