
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,730,055 – 14,730,161 |
| Length | 106 |
| Max. P | 0.864210 |

| Location | 14,730,055 – 14,730,161 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 78.22 |
| Mean single sequence MFE | -46.02 |
| Consensus MFE | -31.18 |
| Energy contribution | -32.35 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
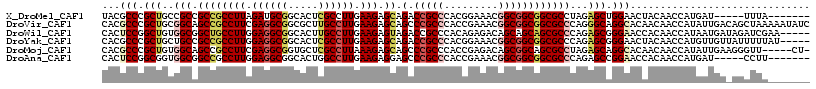
>X_DroMel_CAF1 14730055 106 + 22224390 UACGCCCGCUGCCGCCGCCGCCUUAGAUGCGGCACUCGCCUUGAAGAGCAGACCGCCCACGGAAACGGCGGCGGCGCCUAGAGCUGGAACUACAACCAUGAU-----UUUA------- ....((.((((.((((((((((......(((((.(((........)))..).))))....(....))))))))))).)...))).))...............-----....------- ( -40.10) >DroVir_CAF1 49237 118 + 1 CACGCCCGCUGCGGCAGCCGCCUUCGAGGCGGCGCUUGCCUUGAAGAGCAGCCCGCCCACCGAAACGGCGGCGGCGCCCAGGGCAGGCACAACAACCAUAUUGACAGCUAAAAAUAUC .......(((((((..((((((.....))))))((((((((((..(.((.(((.(((..(((...))).)))))))))))))))))))............))).)))).......... ( -50.30) >DroWil_CAF1 19395 113 + 1 CACUCCGGCUGUGGCGGCUGCCUUGGAGGCGGCACUUGCCUUGAAGAGUAGACCGCCCACAGAGACAGCAGCAGCGCCCAGAGCGGGAACCACAACCAUAAUGAUAGAUCGAA----- (.(((.((((((((((((((((((.((((((.....)))))).))).)))).))).)))))).....((....))..)).))).)((........))................----- ( -39.30) >DroYak_CAF1 22772 113 + 1 CACGCCCGCUGCUGCCGCCGCCUUGGAGGCGGCACUCGCCUUGAAGAGCAGACCGCCCACGGAAACGGCGGCGGCGCCCAGAGCGGGAACUACAACCAUGUUGUUAUUUUUAU----- ....((((((((.(((((((((..(..((((((.(((........)))..).)))))..)(....))))))))))))....))))))(((.(((....))).)))........----- ( -52.30) >DroMoj_CAF1 45460 112 + 1 CACGCCCGCUGUGGCAGCCGCCUUCGAGGCGGUGCUCGCCUUAAAGAGCAGCCCGCCCACCGAGACAGCGGCAGCGCCUAGAGCAGGCACAACAACCAUAUUGAAGGGUU-----CU- ...(((.(((.((((.(((((.((((.((((((((((........)))))..)))))...))))...)))))...))).).))).))).....((((.........))))-----..- ( -45.30) >DroAna_CAF1 21094 106 + 1 CACUCCGGCGGUGGCGGCCGCCUUGGAGGCGGCACUGGCCUUGAAGAGGAGCCCGCCCACCGAAACGGCGGCGGCGCCCAGAGCCGGAACCACAACCAUGAU-----CCUU------- ...(((((((((((((((((((.....))))))...((((((....))).))))).))))).....((((....))))....))))))..............-----....------- ( -48.80) >consensus CACGCCCGCUGCGGCAGCCGCCUUGGAGGCGGCACUCGCCUUGAAGAGCAGACCGCCCACCGAAACGGCGGCGGCGCCCAGAGCAGGAACAACAACCAUAAUGA_AGCUUA_______ ...(((.(((..(((.((((((((.((((((.....)))))).))).)).(.(((((.........))))))))))))...))).))).............................. (-31.18 = -32.35 + 1.17)

| Location | 14,730,055 – 14,730,161 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.22 |
| Mean single sequence MFE | -50.98 |
| Consensus MFE | -35.99 |
| Energy contribution | -36.10 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14730055 106 - 22224390 -------UAAA-----AUCAUGGUUGUAGUUCCAGCUCUAGGCGCCGCCGCCGUUUCCGUGGGCGGUCUGCUCUUCAAGGCGAGUGCCGCAUCUAAGGCGGCGGCGGCAGCGGGCGUA -------....-----....(((........)))((((...(((((((((((((((..(((((((.((.(((......))))).)))).)))...))))))))))))).))))))... ( -49.60) >DroVir_CAF1 49237 118 - 1 GAUAUUUUUAGCUGUCAAUAUGGUUGUUGUGCCUGCCCUGGGCGCCGCCGCCGUUUCGGUGGGCGGGCUGCUCUUCAAGGCAAGCGCCGCCUCGAAGGCGGCUGCCGCAGCGGGCGUG ........(((((((....)))))))....((((((..(((((((((((((((...))))((((((((((((......))).))).))))))....)))))).))).))))))))... ( -54.90) >DroWil_CAF1 19395 113 - 1 -----UUCGAUCUAUCAUUAUGGUUGUGGUUCCCGCUCUGGGCGCUGCUGCUGUCUCUGUGGGCGGUCUACUCUUCAAGGCAAGUGCCGCCUCCAAGGCAGCCGCCACAGCCGGAGUG -----..........((((.((((((((((.(((.....))).)).((.(((((((.((..((((((..(((..(....)..)))))))))..))))))))).)))))))))).)))) ( -46.20) >DroYak_CAF1 22772 113 - 1 -----AUAAAAAUAACAACAUGGUUGUAGUUCCCGCUCUGGGCGCCGCCGCCGUUUCCGUGGGCGGUCUGCUCUUCAAGGCGAGUGCCGCCUCCAAGGCGGCGGCAGCAGCGGGCGUG -----.........(((((...)))))....((((((....(((((((((((......(..((((((..((((........))))))))))..)..))))))))).)))))))).... ( -52.30) >DroMoj_CAF1 45460 112 - 1 -AG-----AACCCUUCAAUAUGGUUGUUGUGCCUGCUCUAGGCGCUGCCGCUGUCUCGGUGGGCGGGCUGCUCUUUAAGGCGAGCACCGCCUCGAAGGCGGCUGCCACAGCGGGCGUG -..-----.......(((((....))))).(((((((...((((..((((((...(((..((((((((((((......))).))).))))))))).))))))))))..)))))))... ( -52.00) >DroAna_CAF1 21094 106 - 1 -------AAGG-----AUCAUGGUUGUGGUUCCGGCUCUGGGCGCCGCCGCCGUUUCGGUGGGCGGGCUCCUCUUCAAGGCCAGUGCCGCCUCCAAGGCGGCCGCCACCGCCGGAGUG -------..((-----(((((....)))))))((((.((((((.(((((((((...)))).))))))).(((.....))))))).)))).((((..(((((......))))))))).. ( -50.90) >consensus _______AAAACU_UCAACAUGGUUGUAGUUCCCGCUCUGGGCGCCGCCGCCGUUUCGGUGGGCGGGCUGCUCUUCAAGGCGAGUGCCGCCUCCAAGGCGGCCGCCACAGCGGGCGUG .............................((((((((...(((((((((.(((((......)))))...........((((.......))))....)))))).)))..)))))))).. (-35.99 = -36.10 + 0.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:14 2006