
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,717,957 – 14,718,048 |
| Length | 91 |
| Max. P | 0.864591 |

| Location | 14,717,957 – 14,718,048 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 79.82 |
| Mean single sequence MFE | -24.92 |
| Consensus MFE | -17.36 |
| Energy contribution | -17.37 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14717957 91 + 22224390 CAUGUUGCCGGCUUCCGACAACCAGGCCAAAAGUUAGGCUAAAAAGCCGACUUUGAGCUUGACAUUGCACUGGGAAAAA---UGGUA----CACC--AAA ..(((.((((..((((......(((((..((((((.((((....))))))))))..))))).((......))))))...---)))))----))..--... ( -26.30) >DroSec_CAF1 11356 88 + 1 CAUGUUGCCGGCUUCCGACAACCAGGCCAAAAGUUAGGCUAAAAAGCCAACUUUAAGCUUGACAUUGCACUGGAAAAAAAGU-GGUA----GG------- ...((((.(((...))).))))(((((..((((((.((((....))))))))))..)))))......((((........)))-)...----..------- ( -26.00) >DroSim_CAF1 11667 94 + 1 CAUGUUGCCGGCUUCCGACAACCAGGCCAAAAGUUAGGCUAAAAAGCCGACUUAAAGCUUGACAUUGCACUGAAAAAAAAAUUGGUA----CACC--AAA ...((((.(((...))).))))(((((...(((((.((((....)))))))))...)))))....................((((..----..))--)). ( -23.50) >DroEre_CAF1 8344 91 + 1 CAUGUUGCCGGCUUCCGACAACCAGGCCAAAAGUUAGGCUAAAGAGCCGAUUUCGAGCUUGACAUUGCACUGGGAACAA---UGGUA----CACC--AAA ..((((.(((((((.........)))))....((((((((...(((.....))).))))))))........)).)))).---(((..----..))--).. ( -25.60) >DroYak_CAF1 10707 91 + 1 CAUGUUGCCGGCUUCCGACAACCAGGCCAAAAGUUAGGCUUACGAGCCGACUUUGAGCUUGACAUUACACUGUGAAAAA---UAGUA----CAUC--AAA ...((((.(((...))).))))(((((..((((((.((((....))))))))))..))))).......(((((.....)---)))).----....--... ( -26.00) >DroAna_CAF1 9042 97 + 1 CGCUUUGCCGGCUUCCGACAAGCGGGCCAAAUCUUGAGCCAAAAUGCAGAUUUAAAGCUUGACAUACCACUGAGAGAAA---UAGGGGGUUUUCCGUGUA .((((((((.((((.....)))).))).((((((...((......)))))))))))))...((((.((.(((.......---)))))((....)))))). ( -22.10) >consensus CAUGUUGCCGGCUUCCGACAACCAGGCCAAAAGUUAGGCUAAAAAGCCGACUUUAAGCUUGACAUUGCACUGGGAAAAA___UGGUA____CACC__AAA ...((((.(((...))).))))(((((...(((((.((((....)))))))))...)))))....................................... (-17.36 = -17.37 + 0.00)

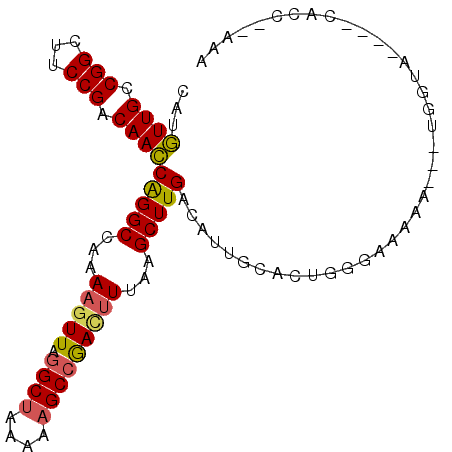
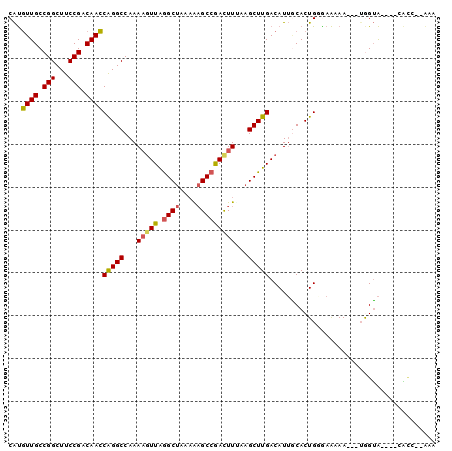
| Location | 14,717,957 – 14,718,048 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 79.82 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -17.55 |
| Energy contribution | -18.97 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.614321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14717957 91 - 22224390 UUU--GGUG----UACCA---UUUUUCCCAGUGCAAUGUCAAGCUCAAAGUCGGCUUUUUAGCCUAACUUUUGGCCUGGUUGUCGGAAGCCGGCAACAUG ...--..((----(((..---.........)))))....((.(((.(((((.((((....))))..))))).))).))((((((((...))))))))... ( -26.60) >DroSec_CAF1 11356 88 - 1 -------CC----UACC-ACUUUUUUUCCAGUGCAAUGUCAAGCUUAAAGUUGGCUUUUUAGCCUAACUUUUGGCCUGGUUGUCGGAAGCCGGCAACAUG -------..----...(-(((........))))......((.(((.((((((((((....)))).)))))).))).))((((((((...))))))))... ( -28.00) >DroSim_CAF1 11667 94 - 1 UUU--GGUG----UACCAAUUUUUUUUUCAGUGCAAUGUCAAGCUUUAAGUCGGCUUUUUAGCCUAACUUUUGGCCUGGUUGUCGGAAGCCGGCAACAUG .((--((..----..))))....................((.(((..((((.((((....))))..))))..))).))((((((((...))))))))... ( -26.50) >DroEre_CAF1 8344 91 - 1 UUU--GGUG----UACCA---UUGUUCCCAGUGCAAUGUCAAGCUCGAAAUCGGCUCUUUAGCCUAACUUUUGGCCUGGUUGUCGGAAGCCGGCAACAUG .((--((..----(..((---(((....)))))..)..))))((.(((((..((((....))))....)))))))...((((((((...))))))))... ( -25.00) >DroYak_CAF1 10707 91 - 1 UUU--GAUG----UACUA---UUUUUCACAGUGUAAUGUCAAGCUCAAAGUCGGCUCGUAAGCCUAACUUUUGGCCUGGUUGUCGGAAGCCGGCAACAUG .((--((((----(((..---...........)).)))))))(((.(((((.((((....))))..))))).)))...((((((((...))))))))... ( -25.22) >DroAna_CAF1 9042 97 - 1 UACACGGAAAACCCCCUA---UUUCUCUCAGUGGUAUGUCAAGCUUUAAAUCUGCAUUUUGGCUCAAGAUUUGGCCCGCUUGUCGGAAGCCGGCAAAGCG ..((((((((........---)))))....))).........(((((((((((((......))...)))))).(((.((((.....)))).)))))))). ( -21.10) >consensus UUU__GGUG____UACCA___UUUUUCCCAGUGCAAUGUCAAGCUCAAAGUCGGCUUUUUAGCCUAACUUUUGGCCUGGUUGUCGGAAGCCGGCAACAUG .......................................((.(((.(((((.((((....))))..))))).))).))((((((((...))))))))... (-17.55 = -18.97 + 1.42)

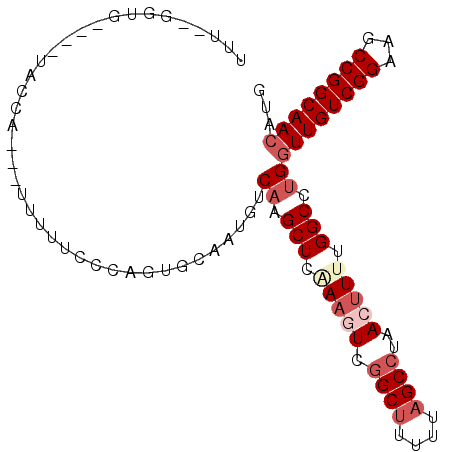
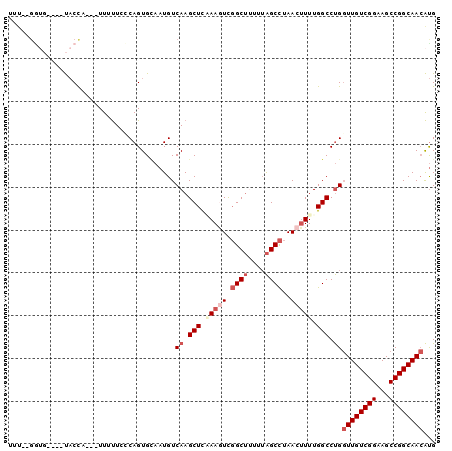
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:08 2006