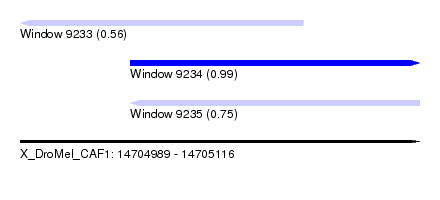
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,704,989 – 14,705,116 |
| Length | 127 |
| Max. P | 0.985323 |

| Location | 14,704,989 – 14,705,079 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 87.88 |
| Mean single sequence MFE | -12.78 |
| Consensus MFE | -8.31 |
| Energy contribution | -8.12 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14704989 90 - 22224390 AGUCAGCGGUUU--GAAAAUCCGACCUAAAUAAUC-CAUAUAUGC----AUACAUAUAUGUACAUAUUUCUCAAACAUCAUCAAAUUCAACCAAUCC .......((((.--(.....).)))).........-((((((((.----...))))))))..................................... ( -10.20) >DroSec_CAF1 23919 90 - 1 AGUCAGCGGUUU--GAAAAUCCGGCCUAAAUAAUC-CAUAUAUGC----AUACAUAUAUGUACAUAUUUUGAAAACAUCAUCAAAUUCAACCAAUCC .......((((.--(((..................-((((((((.----...)))))))).......(((((........))))))))))))..... ( -13.80) >DroEre_CAF1 24294 90 - 1 AGUCAGCGGUUU--GAAAAUCCGAGCUAAAUAAUCAUAUAUAUGC----AUACAUAUAUGUGCAUAUUUCUUAAACAUCGUCAAAUUCAA-CAAUCC .....(((((((--(......))))).(((((..((((((((((.----...))))))))))..))))).........))).........-...... ( -14.20) >DroYak_CAF1 22087 96 - 1 AGUCAGCGGCUUAUAAAAAUCCGACCUAAAUAAUC-CAUAUAUGCCUAUAUACAUAUAUGUACGUAUUUCUUAAACGACGUCAAAUUCAAUCAAUCC .(((..(((.((.....)).)))....(((((...-((((((((........))))))))....))))).......))).................. ( -12.90) >consensus AGUCAGCGGUUU__GAAAAUCCGACCUAAAUAAUC_CAUAUAUGC____AUACAUAUAUGUACAUAUUUCUUAAACAUCAUCAAAUUCAACCAAUCC ......(((...........)))....(((((....((((((((........))))))))....)))))............................ ( -8.31 = -8.12 + -0.19)


| Location | 14,705,024 – 14,705,116 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 79.60 |
| Mean single sequence MFE | -30.00 |
| Consensus MFE | -19.06 |
| Energy contribution | -20.98 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14705024 92 + 22224390 U-------ACAUAUAUGUAU-----GCAUAUAUG-GAUUAU--UUAGGUCGGAUUUUC--AAACCGCUGACUGCGUCUGCGCGGCUGCGUUGCACGUAGUCAACUGAAC .-------.((((((((...-----.))))))))-.....(--(((((.(((......--...))))((((((((((.((((....)))).).))))))))).))))). ( -31.10) >DroGri_CAF1 18399 102 + 1 AAAAAAAAACAGCUAUUUAUUUUGAUCGUAUAUC-GAUUAUCGUUAA-UCGGUCU-----GUGCGGAGCACUGCGUCCGUGCGGUUGCGUUGCACGCUUUCAACUGAAC .........(((.........((((..((..(((-(((((....)))-)))))..-----((((((.((((((((....))))).))).))))))))..)))))))... ( -26.80) >DroSec_CAF1 23954 92 + 1 U-------ACAUAUAUGUAU-----GCAUAUAUG-GAUUAU--UUAGGCCGGAUUUUC--AAACCGCUGACUGCGUCUGCGCGGCUGCGUUGCACGUAGUCAACUGAAC .-------.((((((((...-----.))))))))-.....(--((((((.((......--...))))((((((((((.((((....)))).).))))))))).))))). ( -31.10) >DroEre_CAF1 24328 93 + 1 C-------ACAUAUAUGUAU-----GCAUAUAUAUGAUUAU--UUAGCUCGGAUUUUC--AAACCGCUGACUGCGUCUGCGCGGCUGCGUUGCACGUAGUCAACUGAAC .-------.(((((((((..-----..)))))))))....(--((((..(((......--...))).((((((((((.((((....)))).).))))))))).))))). ( -28.10) >DroYak_CAF1 22122 98 + 1 U-------ACAUAUAUGUAUAUAG-GCAUAUAUG-GAUUAU--UUAGGUCGGAUUUUUAUAAGCCGCUGACUGCGUCUGCGCGGCUGCGUUGCACGUAGUCAACUGAAC .-------.(((((((((......-)))))))))-.....(--(((((.(((.(((....)))))))((((((((((.((((....)))).).))))))))).))))). ( -32.90) >consensus U_______ACAUAUAUGUAU_____GCAUAUAUG_GAUUAU__UUAGGUCGGAUUUUC__AAACCGCUGACUGCGUCUGCGCGGCUGCGUUGCACGUAGUCAACUGAAC .........(((((((((.......))))))))).........((((..(((...........))).((((((((((..(((....)))..).))))))))).)))).. (-19.06 = -20.98 + 1.92)

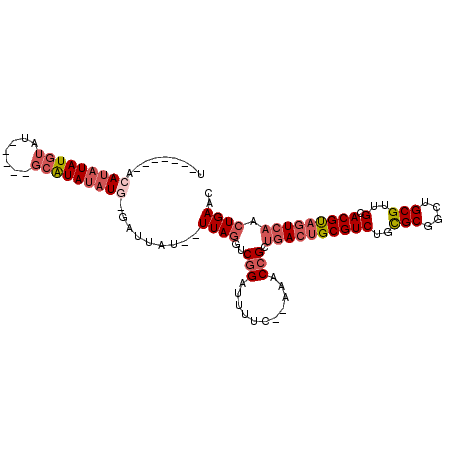
| Location | 14,705,024 – 14,705,116 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 79.60 |
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -13.86 |
| Energy contribution | -15.94 |
| Covariance contribution | 2.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14705024 92 - 22224390 GUUCAGUUGACUACGUGCAACGCAGCCGCGCAGACGCAGUCAGCGGUUU--GAAAAUCCGACCUAA--AUAAUC-CAUAUAUGC-----AUACAUAUAUGU-------A .....(((((((.(((.(..(((....)))..)))).)))))))((((.--(.....).))))...--......-((((((((.-----...)))))))).-------. ( -26.70) >DroGri_CAF1 18399 102 - 1 GUUCAGUUGAAAGCGUGCAACGCAACCGCACGGACGCAGUGCUCCGCAC-----AGACCGA-UUAACGAUAAUC-GAUAUACGAUCAAAAUAAAUAGCUGUUUUUUUUU ...((((((...(((.(((..((..((....))..))..)))..)))..-----....(((-(((....)))))-)..................))))))......... ( -22.10) >DroSec_CAF1 23954 92 - 1 GUUCAGUUGACUACGUGCAACGCAGCCGCGCAGACGCAGUCAGCGGUUU--GAAAAUCCGGCCUAA--AUAAUC-CAUAUAUGC-----AUACAUAUAUGU-------A .....(((((((.(((.(..(((....)))..)))).)))))))(((..--(.....)..)))...--......-((((((((.-----...)))))))).-------. ( -26.50) >DroEre_CAF1 24328 93 - 1 GUUCAGUUGACUACGUGCAACGCAGCCGCGCAGACGCAGUCAGCGGUUU--GAAAAUCCGAGCUAA--AUAAUCAUAUAUAUGC-----AUACAUAUAUGU-------G ((((.(((((((.(((.(..(((....)))..)))).)))))))((..(--....).))))))...--.....((((((((((.-----...)))))))))-------) ( -29.30) >DroYak_CAF1 22122 98 - 1 GUUCAGUUGACUACGUGCAACGCAGCCGCGCAGACGCAGUCAGCGGCUUAUAAAAAUCCGACCUAA--AUAAUC-CAUAUAUGC-CUAUAUACAUAUAUGU-------A .....(((((((.(((.(..(((....)))..)))).)))))))......................--......-((((((((.-.......)))))))).-------. ( -23.00) >consensus GUUCAGUUGACUACGUGCAACGCAGCCGCGCAGACGCAGUCAGCGGUUU__GAAAAUCCGACCUAA__AUAAUC_CAUAUAUGC_____AUACAUAUAUGU_______A .....(((((((.(((.(..(((....)))..)))).)))))))...............................((((((((.........))))))))......... (-13.86 = -15.94 + 2.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:54:03 2006