
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,704,299 – 14,704,421 |
| Length | 122 |
| Max. P | 0.999939 |

| Location | 14,704,299 – 14,704,389 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 75.16 |
| Mean single sequence MFE | -25.83 |
| Consensus MFE | -17.52 |
| Energy contribution | -17.63 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.995039 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14704299 90 + 22224390 AAGAUGUUGCUGCU--------------CUGCGCUGAAGUUAUGCGAAAUGGACGUGGCCGACGUCGCCCACUUCUCAUUUCUUUUAGCCAUCGUUUUUUUUCU-- ..((((..((....--------------..))(((((((.....)((((((((.((((.((....)).)))).)).)))))).))))))))))...........-- ( -25.50) >DroEre_CAF1 23610 98 + 1 AAGAUGUUGUUGAUGCGCAUUGCUGCUGCUGCGCUGAAGUUAUGCGAAAUGGGCGUGGCCGACGUCGCCCACUUCUCAUUUCUUUUUUCCUUUUUUUU-------- ..............(((((..((....))))))).((((..(((.(((.((((((((.....)).)))))).))).)))..)))).............-------- ( -29.00) >DroYak_CAF1 21404 90 + 1 AAGAUGUU----------------GCUACUGCGCUAAAGUUAUGCGAAAUGGGCGUGGCCGACGUCGCCCACUUCUCAUUUCUUUCUUCCAUUUUUUUUUUUGUUU ((((((..----------------((....))...((((..(((.(((.((((((((.....)).)))))).))).)))..))))....))))))........... ( -23.00) >consensus AAGAUGUUG_UG_U__________GCU_CUGCGCUGAAGUUAUGCGAAAUGGGCGUGGCCGACGUCGCCCACUUCUCAUUUCUUUUUUCCAUUUUUUUUUUU_U__ ...................................((((..(((.(((.((((((((.....)).)))))).))).)))..))))..................... (-17.52 = -17.63 + 0.11)



| Location | 14,704,299 – 14,704,389 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 75.16 |
| Mean single sequence MFE | -25.87 |
| Consensus MFE | -17.82 |
| Energy contribution | -17.60 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.69 |
| SVM decision value | 4.69 |
| SVM RNA-class probability | 0.999939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14704299 90 - 22224390 --AGAAAAAAAACGAUGGCUAAAAGAAAUGAGAAGUGGGCGACGUCGGCCACGUCCAUUUCGCAUAACUUCAGCGCAG--------------AGCAGCAACAUCUU --...........(((((((....((((((.((.((((.((....)).)))).))))))))...........((....--------------.)))))..)))).. ( -26.30) >DroEre_CAF1 23610 98 - 1 --------AAAAAAAAGGAAAAAAGAAAUGAGAAGUGGGCGACGUCGGCCACGCCCAUUUCGCAUAACUUCAGCGCAGCAGCAGCAAUGCGCAUCAACAACAUCUU --------.......((((.....((((((.((((((((((..........)))))))))).)))...))).(((((((....))..)))))..........)))) ( -28.80) >DroYak_CAF1 21404 90 - 1 AAACAAAAAAAAAAAUGGAAGAAAGAAAUGAGAAGUGGGCGACGUCGGCCACGCCCAUUUCGCAUAACUUUAGCGCAGUAGC----------------AACAUCUU ..............(((....((((..(((.((((((((((..........)))))))))).)))..)))).((......))----------------..)))... ( -22.50) >consensus __A_AAAAAAAAAAAUGGAAAAAAGAAAUGAGAAGUGGGCGACGUCGGCCACGCCCAUUUCGCAUAACUUCAGCGCAG_AGC__________A_CA_CAACAUCUU ......................(((..(((.((((((((((..........)))))))))).)))..))).................................... (-17.82 = -17.60 + -0.22)



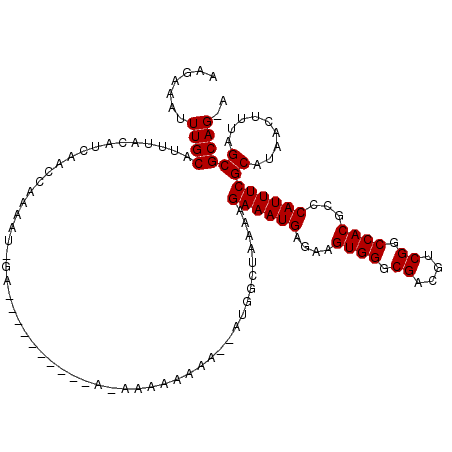
| Location | 14,704,313 – 14,704,421 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.00 |
| Mean single sequence MFE | -25.37 |
| Consensus MFE | -20.46 |
| Energy contribution | -21.47 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.953804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14704313 108 + 22224390 --CUGCGCUGAAGUUAUGCGAAAUGGACGUGGCCGACGUCGCCCACUUCUCAUUUCUUUUAGCCAUCGUUUUUUUUCU----------UCCAUUUUGUUUGAUGUAAAUGCAAAUUUCUU --.((((((((((.....)((((((((.((((.((....)).)))).)).)))))).))))))(((((..........----------...........))))).....)))........ ( -24.70) >DroSec_CAF1 23272 93 + 1 UGCUGCGCUAAAGUUAUGCGAAAUGGGCGUGGCCGACGUCGCCCACUUCUCAUUUCUUUUAGCC---------------------------AUUUUGGUUGAUGUAAAUGCAAAUUUCUU ...((((((((((..(((.(((.((((((((.....)).)))))).))).)))..).))))))(---------------------------(((......)))).....)))........ ( -27.70) >DroYak_CAF1 21414 120 + 1 UACUGCGCUAAAGUUAUGCGAAAUGGGCGUGGCCGACGUCGCCCACUUCUCAUUUCUUUCUUCCAUUUUUUUUUUUGUUUUGUUGUUGUCUAUUUUGGUUGAUGUUAAUGCAAAUUUCUU ...(((...((((..(((.(((.((((((((.....)).)))))).))).)))..))))......................((((..(((..........)))..)))))))........ ( -23.70) >consensus U_CUGCGCUAAAGUUAUGCGAAAUGGGCGUGGCCGACGUCGCCCACUUCUCAUUUCUUUUAGCCAU__UUUUUUUU_U__________UC_AUUUUGGUUGAUGUAAAUGCAAAUUUCUU ...((((((((((..(((.(((.((((((((.....)).)))))).))).)))..)))).)))...........................((((......)))).....)))........ (-20.46 = -21.47 + 1.00)

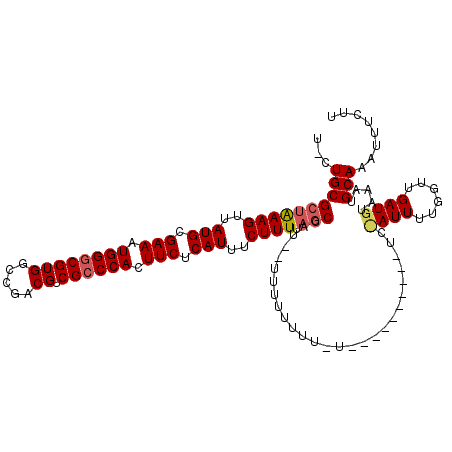

| Location | 14,704,313 – 14,704,421 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.00 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -20.37 |
| Energy contribution | -20.37 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.80 |
| SVM decision value | 3.22 |
| SVM RNA-class probability | 0.998775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14704313 108 - 22224390 AAGAAAUUUGCAUUUACAUCAAACAAAAUGGA----------AGAAAAAAAACGAUGGCUAAAAGAAAUGAGAAGUGGGCGACGUCGGCCACGUCCAUUUCGCAUAACUUCAGCGCAG-- ..(((...(((.....((((............----------...........)))).......((((((.((.((((.((....)).)))).)))))))))))....))).......-- ( -24.90) >DroSec_CAF1 23272 93 - 1 AAGAAAUUUGCAUUUACAUCAACCAAAAU---------------------------GGCUAAAAGAAAUGAGAAGUGGGCGACGUCGGCCACGCCCAUUUCGCAUAACUUUAGCGCAGCA .......((((.....(((........))---------------------------)((((((.(..(((.((((((((((..........)))))))))).)))..))))))))))).. ( -27.50) >DroYak_CAF1 21414 120 - 1 AAGAAAUUUGCAUUAACAUCAACCAAAAUAGACAACAACAAAACAAAAAAAAAAAUGGAAGAAAGAAAUGAGAAGUGGGCGACGUCGGCCACGCCCAUUUCGCAUAACUUUAGCGCAGUA .......((((...........(((..............................)))...((((..(((.((((((((((..........)))))))))).)))..))))...)))).. ( -23.51) >consensus AAGAAAUUUGCAUUUACAUCAACCAAAAU_GA__________A_AAAAAAAA__AUGGCUAAAAGAAAUGAGAAGUGGGCGACGUCGGCCACGCCCAUUUCGCAUAACUUUAGCGCAG_A .......((((.....................................................((((((....((((.((....)).))))...))))))((.........)))))).. (-20.37 = -20.37 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:59 2006