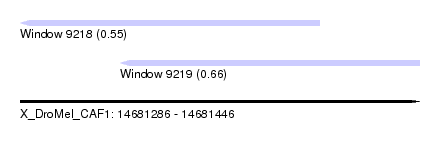
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,681,286 – 14,681,446 |
| Length | 160 |
| Max. P | 0.663808 |

| Location | 14,681,286 – 14,681,406 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.67 |
| Mean single sequence MFE | -54.40 |
| Consensus MFE | -44.14 |
| Energy contribution | -42.82 |
| Covariance contribution | -1.32 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.546683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14681286 120 - 22224390 CGGUGGCGUAGAGACGCUCACCUUCAUCAAGGUGCCGUCGGGCAAGAGUCCGGCCACCCAACCCGAUGUGGAACUCAUCCAAGUGGCCGGCAGCCUGGCUAGCGAUGAUGGCACUGCCCU .((((((((....)))).))))........((((((((((.((....(.((((((((.((......))((((.....)))).)))))))))(((...))).))..))))))))))..... ( -50.00) >DroYak_CAF1 12564 120 - 1 CGGCGGCGUGGAGACGCUCACCUUCAUCAAGGUGCCAUCGGGCAAGAGUCCGGCCAGCCAACCCGAUGUGGAACUCAUCCAGGUGGCCGGCAGCUUGGCCAGCGAUGAUGGCACCGCCCU .(((((.(((....)))...))........(((((((((((((....))))((((((((...((.((.((((.....)))).))))..)))....)))))......)))))))))))).. ( -55.40) >DroAna_CAF1 16709 120 - 1 CGGGGGCGUGGAGACACUGACCUUCAUCAAGGUGCCCUCGGCCCAGAGCCCACCCACCCAGCCGGACAUCGAGCUCAUCCAGGUGGCCGGCAGCCUGGCCAGCGACGAGGGCACCGCCCU ..((((((((....))).............((((((((((.....((((.(.....((.....)).....).)))).....(.(((((((....))))))).)..))))))))))))))) ( -57.80) >consensus CGGCGGCGUGGAGACGCUCACCUUCAUCAAGGUGCCAUCGGGCAAGAGUCCGGCCACCCAACCCGAUGUGGAACUCAUCCAGGUGGCCGGCAGCCUGGCCAGCGAUGAUGGCACCGCCCU .((.((((((....))).............((((((((((.....(((((((................))).)))).....(.(((((((....))))))).)..))))))))))))))) (-44.14 = -42.82 + -1.32)

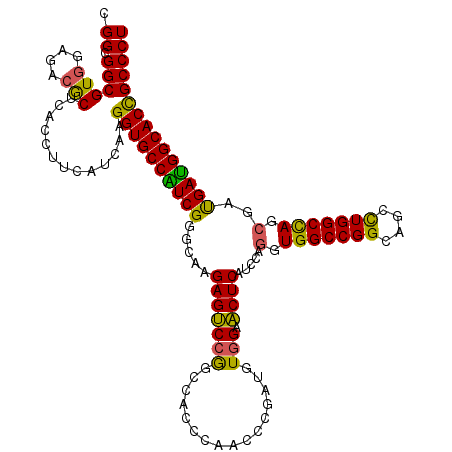
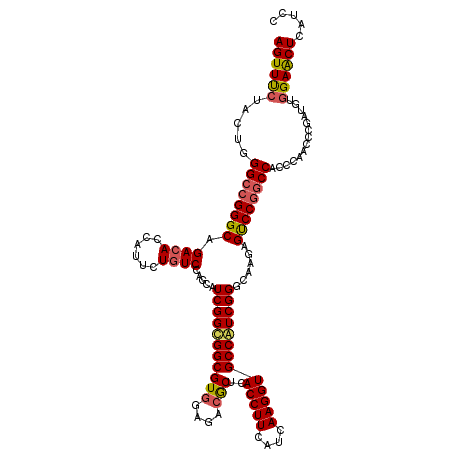
| Location | 14,681,326 – 14,681,446 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.56 |
| Mean single sequence MFE | -49.00 |
| Consensus MFE | -35.84 |
| Energy contribution | -36.74 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14681326 120 - 22224390 AGUUUCUACUGGGCCGGGCAGACACCUUUCUGUCCAGUAUCGGUGGCGUAGAGACGCUCACCUUCAUCAAGGUGCCGUCGGGCAAGAGUCCGGCCACCCAACCCGAUGUGGAACUCAUCC ...((((((((((..(((((((......)))))))......((((((.....((((..((((((....)))))).))))((((....)))).))))))...))))..))))))....... ( -50.00) >DroYak_CAF1 12604 120 - 1 AGUUUCUACUGGGCCGGGCAGACACCAUUCUCUCCAGCAUCGGCGGCGUGGAGACGCUCACCUUCAUCAAGGUGCCAUCGGGCAAGAGUCCGGCCAGCCAACCCGAUGUGGAACUCAUCC ...((((((((((...((((((......)))..........(((((((((....)))..(((((....))))))))..(((((....)))))))).)))..))))..))))))....... ( -42.80) >DroAna_CAF1 16749 120 - 1 AGUUCCUGCUGGGCCGGGCGGACACCAUCAUGUCCAGCAUCGGGGGCGUGGAGACACUGACCUUCAUCAAGGUGCCCUCGGCCCAGAGCCCACCCACCCAGCCGGACAUCGAGCUCAUCC .((.((.((((((..(((((((((......)))))....(((((((((((....)))..(((((....)))))))))))))......)))).....)))))).))))............. ( -54.20) >consensus AGUUUCUACUGGGCCGGGCAGACACCAUUCUGUCCAGCAUCGGCGGCGUGGAGACGCUCACCUUCAUCAAGGUGCCAUCGGGCAAGAGUCCGGCCACCCAACCCGAUGUGGAACUCAUCC ((((((.....((((((((.((((......)))).....(((((((((((....)))..(((((....)))))))))))))......))))))))..............))))))..... (-35.84 = -36.74 + 0.90)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:49 2006