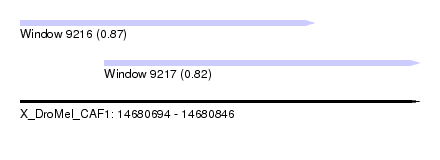
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,680,694 – 14,680,846 |
| Length | 152 |
| Max. P | 0.870284 |

| Location | 14,680,694 – 14,680,806 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.39 |
| Mean single sequence MFE | -46.47 |
| Consensus MFE | -26.01 |
| Energy contribution | -25.47 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870284 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14680694 112 + 22224390 -----UUGGUGAUCUUCAUCUGGA---GUUAAUCAACUCCAUUCGGAUCGUAUCAUAUCAGCCGCCUUUUCCCCAAUCAUAAAUGCCGCUGCAUUUGUGUGAGCGGUGGGUGGAAACGGU -----..((((((((.....((((---(((....)))))))...)))))(........).)))((((((((((((..(((((((((....))))))))).......)))).))))).))) ( -35.00) >DroYak_CAF1 11967 117 + 1 UUCUGUUGAUGAUCGUCAUCUGCC---GUUGAUCAACUCCAUUCGGAUCGUAUCAUAUCCGCCGCCUUCUCCCCAAUCAUGAAGGCCGCUGCAUUCGUGUGGGCGGUGGGCGGGAAUGGG ....((((((.(.((.(....).)---).).)))))).((((((((((........)))).((((((.((.((((..((((((.((....)).)))))))))).)).)))))))))))). ( -46.00) >DroAna_CAF1 16110 119 + 1 CUGUUUUGGAAACCCA-AACCGGAGCCGGCCCUCAGCUCCAGUCGGCCCGCACCAGAUCCGCCGCCUUCUCGCCGAUCAUGAAGGCCGCCGCAUUGGUGUGGGCGGUGGGCGGCACCGGG ....(((((....)))-))((((.((((.(((.(.((....))..(((((((((((..(.((.((((((...........)))))).)).)..))))))))))).).))))))).)))). ( -58.40) >consensus _U_U_UUGGUGAUCCUCAUCUGGA___GUUAAUCAACUCCAUUCGGAUCGUAUCAUAUCCGCCGCCUUCUCCCCAAUCAUGAAGGCCGCUGCAUUCGUGUGGGCGGUGGGCGGAAACGGG ...................(((.............(((((....)))..))......((((((((((((...........))))))((((((..........))))))))))))..))). (-26.01 = -25.47 + -0.54)


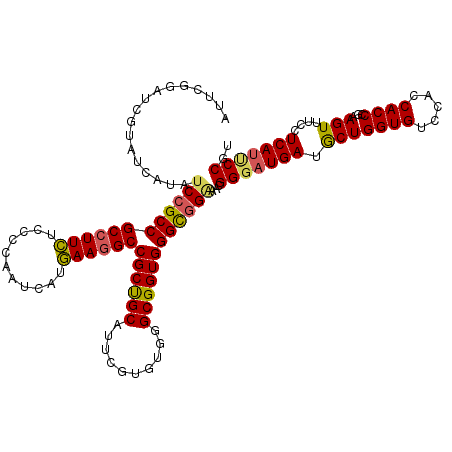
| Location | 14,680,726 – 14,680,846 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.17 |
| Mean single sequence MFE | -49.90 |
| Consensus MFE | -35.29 |
| Energy contribution | -35.63 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 14680726 120 + 22224390 AUUCGGAUCGUAUCAUAUCAGCCGCCUUUUCCCCAAUCAUAAAUGCCGCUGCAUUUGUGUGAGCGGUGGGUGGAAACGGUAUGAUACUGGUGUCCACCACCCGAAGUUUCCUCACUCCAU .(((((...((((((((((..((((((...((.(...(((((((((....)))))))))...).)).))))))....))))))))))((((....)))).)))))............... ( -43.90) >DroYak_CAF1 12004 120 + 1 AUUCGGAUCGUAUCAUAUCCGCCGCCUUCUCCCCAAUCAUGAAGGCCGCUGCAUUCGUGUGGGCGGUGGGCGGGAAUGGGAUGAUGCUGGUGUCCACCACCCGGAGUUUCCUCAUUCCGU ....((...(((((((.((((((((((.((.((((..((((((.((....)).)))))))))).)).))))))...)))))))))))((((....))))))((((((......)))))). ( -47.60) >DroAna_CAF1 16149 120 + 1 AGUCGGCCCGCACCAGAUCCGCCGCCUUCUCGCCGAUCAUGAAGGCCGCCGCAUUGGUGUGGGCGGUGGGCGGCACCGGGAUGAUGCUGGUGUCCACCACCCGCAGUCGCCUCAUCCCGU .....(((((((((((..(.((.((((((...........)))))).)).)..)))))))))))((((.....))))((((((((((.((((.....)))).)))......))))))).. ( -58.20) >consensus AUUCGGAUCGUAUCAUAUCCGCCGCCUUCUCCCCAAUCAUGAAGGCCGCUGCAUUCGUGUGGGCGGUGGGCGGAAACGGGAUGAUGCUGGUGUCCACCACCCGAAGUUUCCUCAUUCCGU .................((((((((((((...........))))))((((((..........))))))))))))...(((((((.(((((((.....))))...)))....))))))).. (-35.29 = -35.63 + 0.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:47 2006