| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 14,678,778 – 14,678,875 |
| Length | 97 |
| Max. P | 0.783420 |

| Location | 14,678,778 – 14,678,875 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 90.72 |
| Mean single sequence MFE | -34.10 |
| Consensus MFE | -27.76 |
| Energy contribution | -28.07 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
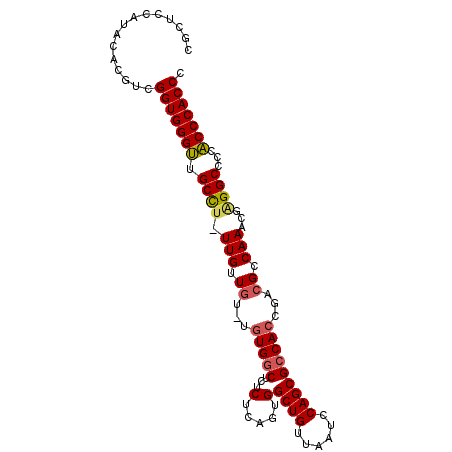
>X_DroMel_CAF1 14678778 97 + 22224390 CGUUCCAUACACGUCGGUGGGUUGCUU-UUGUUUUCUGUGGCUCUCUCAGUGGCUGUUAAUCCAGCGCCACCGAUGCCAAACGAGGCCCCACCCACCC ...............(((((((.((((-(.(((.....((((....((.(((((.(((.....)))))))).)).))))))))))))...))))))). ( -32.10) >DroSec_CAF1 11553 96 + 1 CGUUCCAUACAUGUCGGUGGGUUGCCU-UUGUUGU-UGUGGCUCUCUCAGUGGCUGUUAAUCCAGCGCCACCGACGCCAAACGCGGCCCCACCCACCC ............((.((((((((((.(-(((.(((-((((((...(.....)((((......)))))))).))))).)))).)))).)))))).)).. ( -34.90) >DroEre_CAF1 12662 96 + 1 CGCUUCAUACACGUCGGUGGGCUGCCU-UUGUUGU-UGUGGCUCUCUCAGUGGCUGUUAAUCCAGCGCCACCGCCGCCAAAUGGGGCCCCGCCCACCC ...............(((((((.((((-(..(((.-.(((((.......(((((.(((.....)))))))).))))))))..)))))...))))))). ( -35.70) >DroYak_CAF1 10443 97 + 1 CGCUCCAUACACGUCGGUGGGUUGCCUUUUGUUGU-UGUGACGCUCUCAGUGGCUGUUAAUCCAGCGCCAACGUCGCCAAAUGAGGCCCCACCCACCC ...............(((((((.(((((...(((.-.((((((.......((((.(((.....))))))).)))))))))..)))))...))))))). ( -33.70) >consensus CGCUCCAUACACGUCGGUGGGUUGCCU_UUGUUGU_UGUGGCUCUCUCAGUGGCUGUUAAUCCAGCGCCACCGACGCCAAACGAGGCCCCACCCACCC ...............(((((((.((((.(((.((...(((((...(.....)((((......)))))))))...)).)))...))))...))))))). (-27.76 = -28.07 + 0.31)


| Location | 14,678,778 – 14,678,875 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 90.72 |
| Mean single sequence MFE | -37.12 |
| Consensus MFE | -33.01 |
| Energy contribution | -32.95 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.783420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
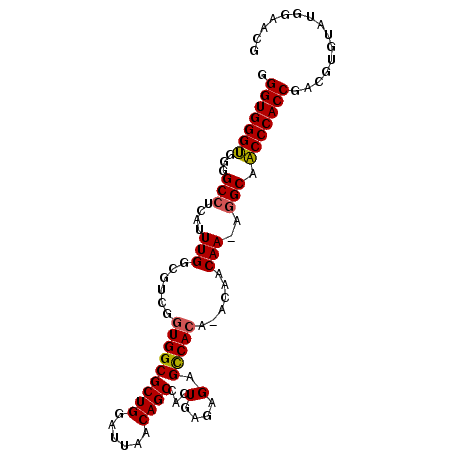
>X_DroMel_CAF1 14678778 97 - 22224390 GGGUGGGUGGGGCCUCGUUUGGCAUCGGUGGCGCUGGAUUAACAGCCACUGAGAGAGCCACAGAAAACAA-AAGCAACCCACCGACGUGUAUGGAACG ..((.(((((((((......))).((.(((((((((......))))...(....).))))).))......-......)))))).))............ ( -36.00) >DroSec_CAF1 11553 96 - 1 GGGUGGGUGGGGCCGCGUUUGGCGUCGGUGGCGCUGGAUUAACAGCCACUGAGAGAGCCACA-ACAACAA-AGGCAACCCACCGACAUGUAUGGAACG .(((((((...(((..(((((((.((((((((..((......))))))))))....))))..-..)))..-.))).)))))))............... ( -39.50) >DroEre_CAF1 12662 96 - 1 GGGUGGGCGGGGCCCCAUUUGGCGGCGGUGGCGCUGGAUUAACAGCCACUGAGAGAGCCACA-ACAACAA-AGGCAGCCCACCGACGUGUAUGAAGCG .(((((((...(((...((((...(..(((((((((......))))...(....).))))).-.)..)))-)))).)))))))..(((.......))) ( -38.70) >DroYak_CAF1 10443 97 - 1 GGGUGGGUGGGGCCUCAUUUGGCGACGUUGGCGCUGGAUUAACAGCCACUGAGAGCGUCACA-ACAACAAAAGGCAACCCACCGACGUGUAUGGAGCG ..((.(((((((((......)))((((((..(((((......))))....)..))))))...-..............)))))).))............ ( -34.30) >consensus GGGUGGGUGGGGCCUCAUUUGGCGUCGGUGGCGCUGGAUUAACAGCCACUGAGAGAGCCACA_ACAACAA_AGGCAACCCACCGACGUGUAUGGAACG .(((((((...(((....(((......(((((((((......))))...(....).)))))......)))..))).)))))))............... (-33.01 = -32.95 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:53:45 2006